Mitigating Bias in Microbiome Workflows through the Implementation of Microbiome Standards and Controls
The Human Microbiome Project Phase 1 (2007-2012) sparked significant interest in microbiome research, particularly in the realm of the human gut microbiome. Due to the novelty of this field and the incomplete validation of wet-lab and dry-lab protocols, coupled with the intricate and bias-prone nature of Next-Gen Sequencing workflows, researchers grappled with the pervasive issue of data quality in publications. The challenge intensified when comparing data across laboratories, as highlighted in a notable 2014 Science News publication. This publication showcased substantial discrepancies in profiling results for the same stool sample processed by two renowned organizations (Figure 1).
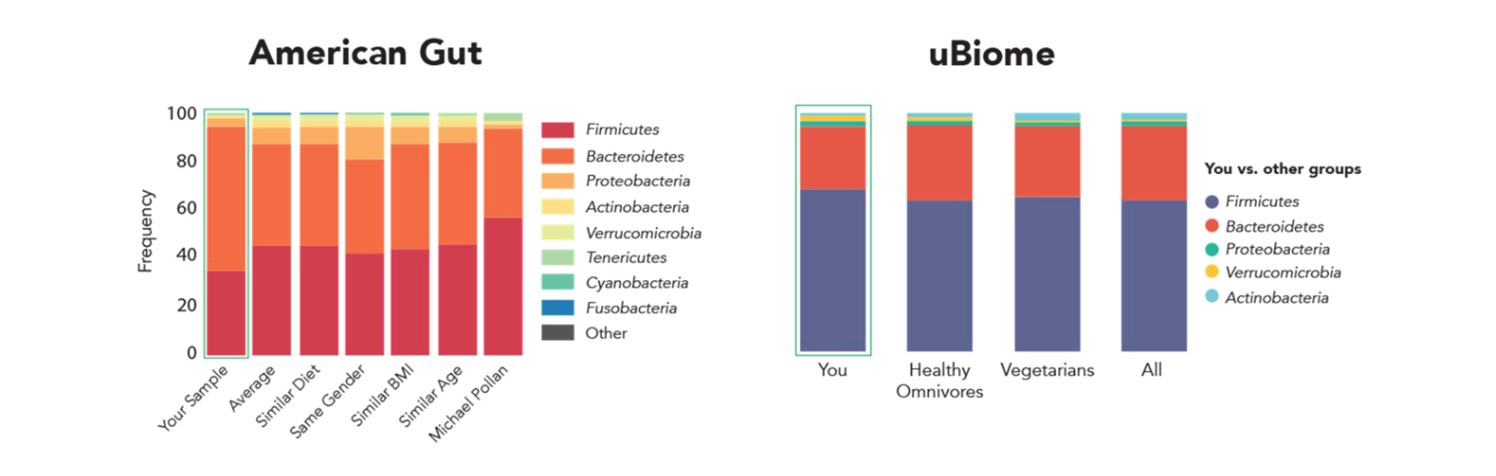
Figure 1. American Gut and uBiome report contradictory results from the same fecal sample, demonstrating the lack of reproducibility in microbiome research.
Recognizing the need for microbiome standards and controls to address bias in this emerging field, Zymo Research and other key players such as the National Institute of Standards and Technology (NIST) and American Type Culture Collection (ATCC) took proactive steps. Beginning in 2014, two years were dedicated to the development of the first microbiome standard: the ZymoBIOMICS Microbial Community Standard. Zymo Research entered into competition with ATCC when introducing the product at a NIST workshop in 2016, and successfully launched the first commercial microbiome standard in 2017. This launch coincided with the introduction of the first microbial DNA extraction kit validated for unbiased microbiome profiling, in the form of the ZymoBIOMICS DNA Miniprep Kit. Today, Zymo Research’s microbiome standards have garnered approximately nine to ten times more citations than the competition (Figure 2).
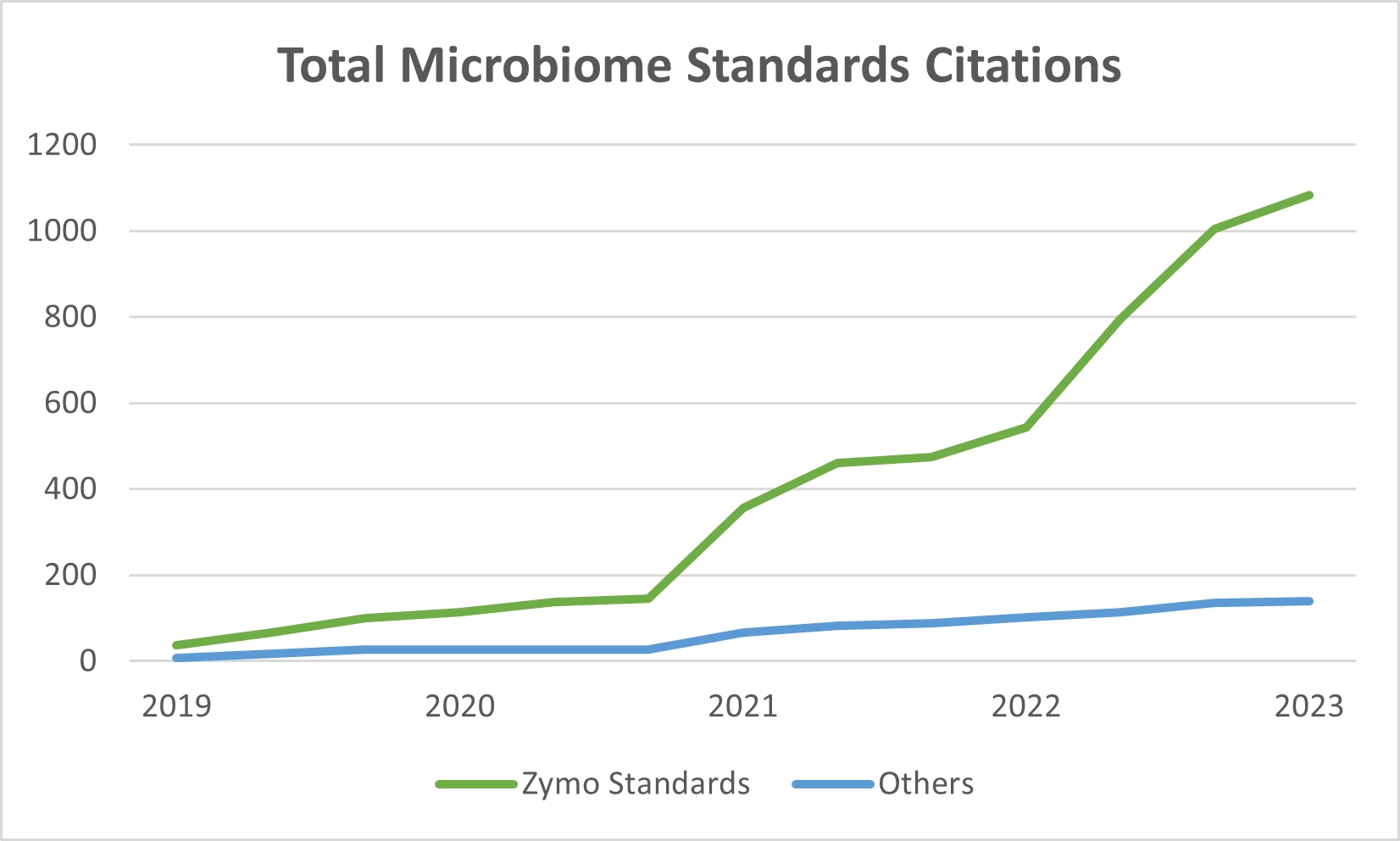
Figure 2. Zymo Research’s microbiome standards have been cited 1,083 times, while all other competing standards have been cited a total of 140 times.
This success has been hard-won; however, initially, our microbiome standards faced a mixed reception in the market. Resistance did not arise from inherent quality issues, but instead from challenges arising from measuring and comparing bias in established workflows. The previous gold standard of workflows, the fecal microbiome profiling workflow employed by the Human Microbiome Project, was revealed to be among the most biased. The primary source of bias was identified in the PowerSoil DNA Isolation Kit, which utilized 0.7mm Garnet beads for mechanical lysis. Internal research demonstrated that these beads were too large for effective bacterial lysis. As such, measuring the potential impact of bias in widely used workflows has become an important step in assuring the integrity of current and future microbiome studies.
Assessing bias at scale remains a complex challenge. Zymo Research continues to extend support to researchers through various channels such as conference talks, blogs, emails, and publications, but more support is required to sustain this rapidly growing field. To extend the reach of these efforts, Zymo Research created the Microbiome Standards and Controls Initiative (M-SCI). Through M-SCI, thousands of microbiome standards have been provided to labs across the world, free of charge. This initiative has encouraged researchers to incorporate controls into their workflows, fostering a deeper understanding of bias and increasing overall data quality (Figure 3).
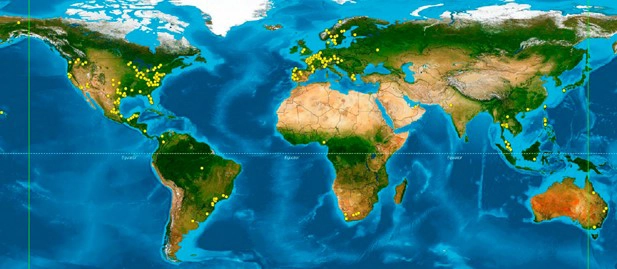
Figure 3. Over 460 participating labs across 47 countries have joined in the M-SCI initiative.
M-SCI has been warmly received. In a recent microbiome standard workshop hosted by NIST, researchers from organizations across the United States actively engaged in discussions surrounding bias — a term that is no longer taboo. By continuing to create and foster bias mitigation strategies and create spaces for open discussion, Zymo Research hopes to help usher in the best future for the microbiome field.