A Global Leader in Wastewater Surveillance



A Global Leader in Wastewater Surveillance
Zymo Research is a trusted leader in wastewater-based epidemiology, partnering with top public health institutions to advance infectious disease monitoring and outbreak prevention. With deep expertise in sample collection and stabilization, nucleic acid purification, and both PCR- and NGS-based molecular analysis, we offer a complete toolkit for high-resolution pathogen detection and antimicrobial resistance tracking. Whether you're building in-house capabilities or seeking a fully outsourced solution, our technologies and comprehensive services are designed to deliver accurate, actionable insights. By working closely with clinical, academic, and public health experts, we help communities stay ahead of emerging health threats, turning wastewater into a powerful tool for protecting global health.
A New Frontier in Public Health Monitoring
Wastewater surveillance provides a non-invasive snapshot of circulating pathogens and antibiotic resistance genes, supporting genomic epidemiology at scale. Discover how the field is delivering high-resolution, real-time insights into infectious disease dynamics and strengthening public health infrastructure. Read more
Wastewater Monitoring in 4 Simple Steps
Detect and Monitor Pathogens with Unparalleled Sensitivity
Step 1: Sample Collection & Preservation
Wastewater Stabilization Buffer & Collection Device
Ideal for the storage, preservation, and concentration of liquid samples including water, wastewater, sewage, finished water, natural water, river water, fresh water, salt water, etc.



Wastewater Stabilization Buffer (WSB) is a specialized preservation solution designed to maintain DNA and RNA integrity in various water samples, including wastewater, sewage, and natural water sources. WSB effectively inactivates pathogens while preserving nucleic acids and microbial composition for up to a week at room temperature, eliminating the need for refrigeration or cold-chain logistics. This specialized buffer removes the need for ultrafiltration or ultracentrifugation, simplifying the concentration of large liquid samples for molecular testing. The Wastewater Sample Collection Bottles are prefilled with WSB, offering a convenient, all-in-one solution for the safe collection, transport, and storage of wastewater samples. Optimized for wastewater-based epidemiology, public health monitoring, and environmental research, these technologies provide a dependable and efficient approach to wastewater sample collection, preservation, and analysis.
Sample Stability for Accurate Results
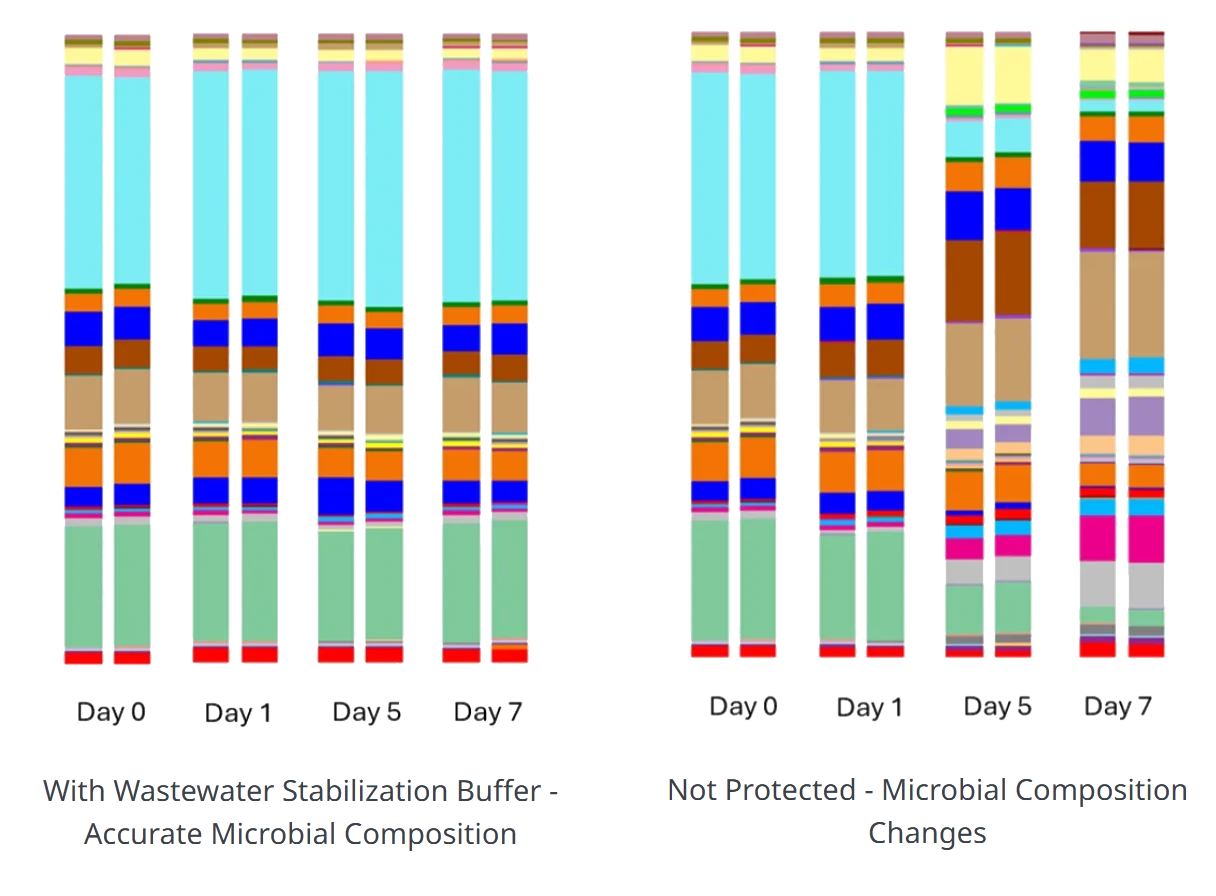
Comparison of microbial composition in wastewater samples over time with and without Wastewater Stabilization Buffer (WSB). The left panel demonstrates that using WSB preserves microbial composition accurately from Day 0 to Day 7. In contrast, the right panel shows that in the absence of stabilization, microbial composition shifts significantly over time, leading to potential inaccuracies in wastewater-based epidemiology studies.
DNA/RNA Shield™
Ideal for the storage, preservation, and processing of solid samples including water filters, wastewater solids, sludge, concentrated water samples, etc.



DNA/RNA Shield™ preserves nucleic acids at ambient temperatures from the moment of sample collection, eliminating the need for cold-chain shipping and reducing transport costs. DNA remains stable for at least two years, while RNA is preserved for up to 30 days.
Solid wastewater samples collected and homogenized in DNA/RNA Shield™ can be seamlessly transferred to lysis and nucleic acid purification workflows. Additionally, the buffer completely inactivates pathogens including viruses, bacteria, fungi, and parasites, ensuring safe handling. The stabilization solution also protects against degradation from freeze-thaw cycling and unexpected freezer failures, maintaining sample integrity for reliable analysis.
Preservation of Genetic Integrity at Ambient Temperatures
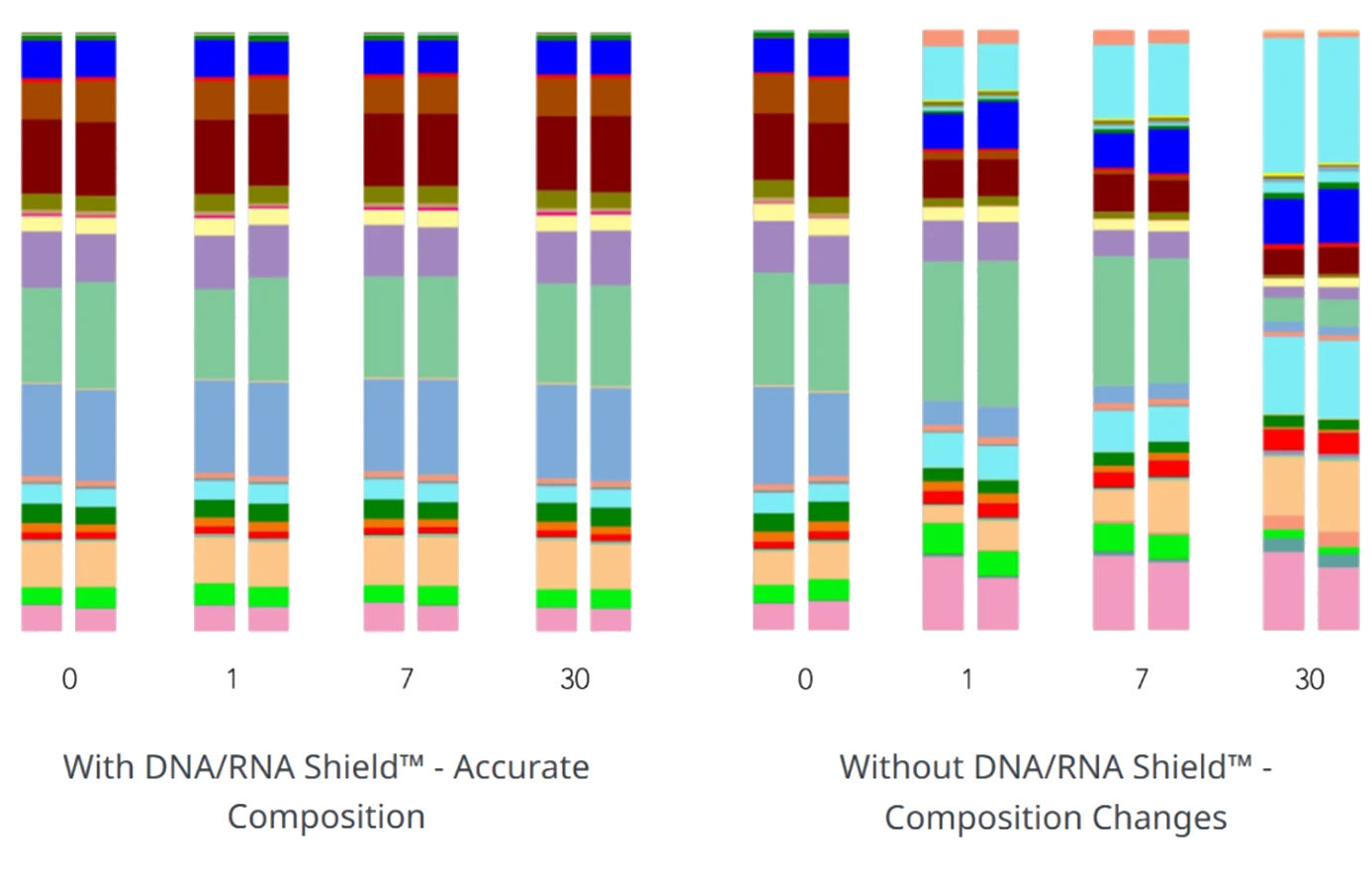
Microbial composition of stool remains unchanged after one month at ambient temperature when collected and stored in DNA/RNA Shield™. Extracted DNA was analyzed via 16S rRNA gene sequencing, confirming that DNA/RNA Shield™ preserves genetic integrity at the point of collection and prevents nucleic acid degradation. This makes it ideal for workflows requiring sensitive downstream analyses, such as Next-Gen Sequencing and RT-PCR.
Effective Inactivation of Viruses, Bacteria, and Fungi
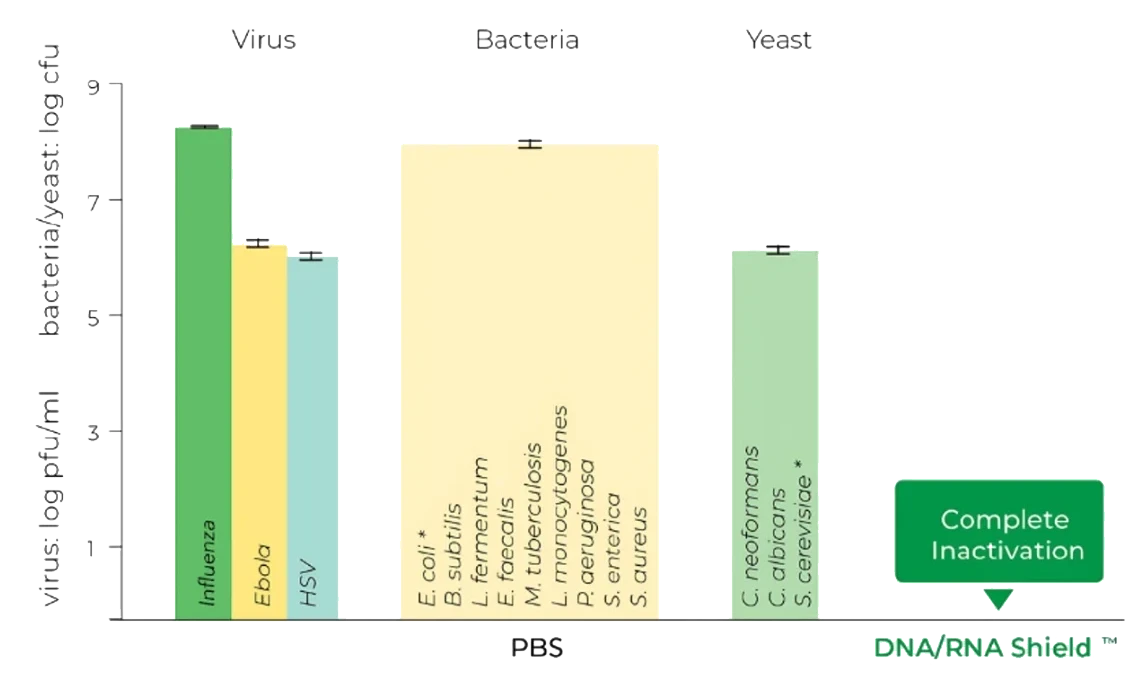
Viruses, bacteria, and yeast are inactivated by DNA/RNA Shield™. Samples containing infectious agents were treated for 5 minutes with DNA/RNA Shield™ or PBS. Titer (PFU) was subsequently determined by plaque assay. These results were validated by D. Poole and Prof. A Mehle, Department of Medical Microbiology and Immunology at the University of Wisconsin, Madison (Influenza A-D); L. Avena and Dr. A Griffiths, Department of Virology and Immunology at Texas Biomedical Research Institute (Ebola); H. Oh, F. Diaz, and Prof. D. Knipe, Virology Program at Harvard Medical School (HSV-1/2); and Zymo Research Corporation (E. coli, L. fermentum, B. subtilis, and S. cerevisiae).
*Disclaimer: This graph only displays results from E. coli inactivation. Each microbe was tested independently and were combined into one graph for brevity. Bacterial cultures were grown between 108 – 109 cells and yeast cultures were grown between 107 – 108 cells.
Step 2: DNA/RNA Extraction
Quick-DNA/RNA™ Water Kit
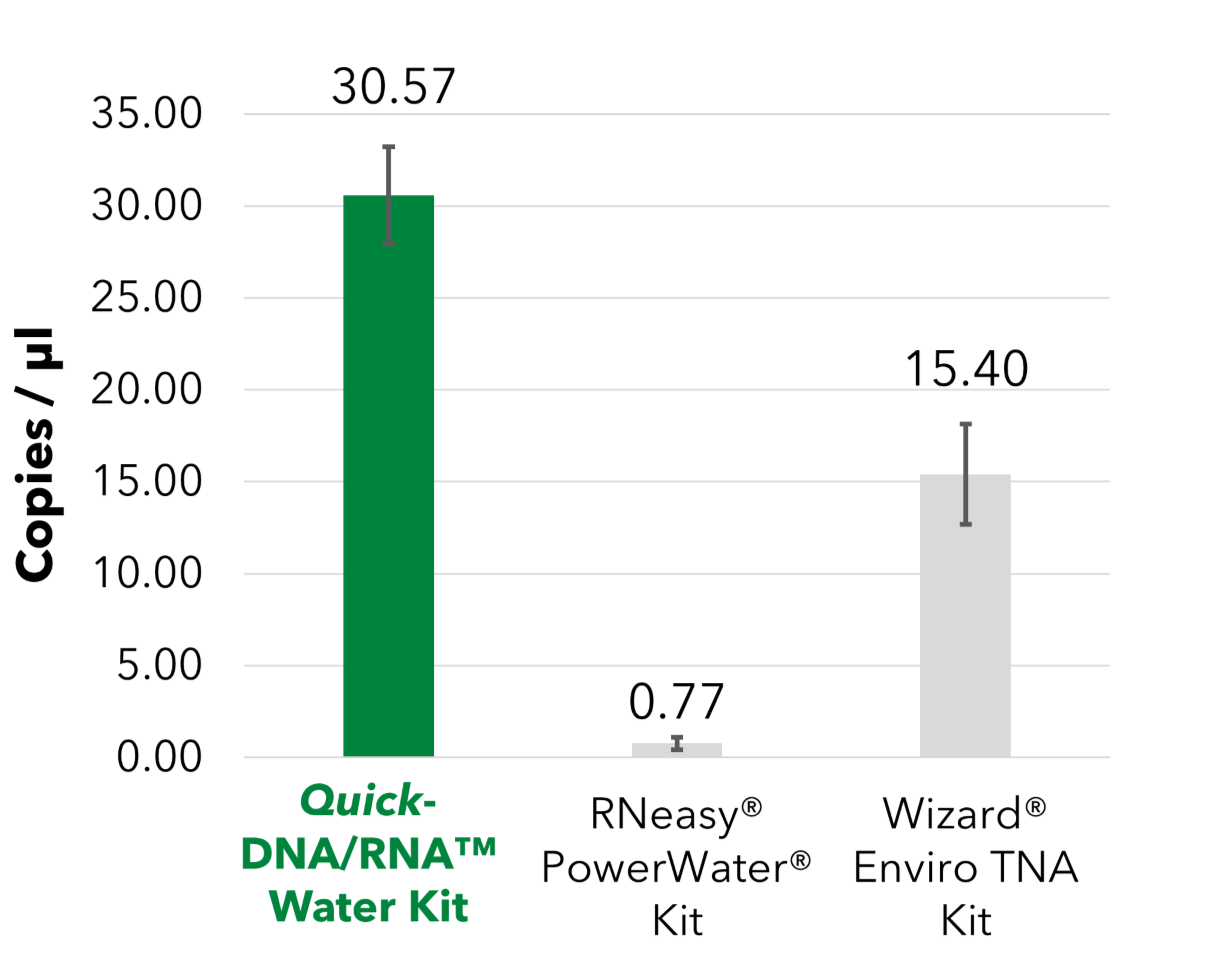
The Quick-DNA/RNA™ Water Kit enables efficient purification of inhibitor-free nucleic acids from large water volumes. Designed for total nucleic acid extraction from viruses, bacteria, fungi, and other waterborne pathogens, it supports a wide range of sample types, including low-biomass water, raw wastewater, sewage, sludge, treated water, natural water, rivers, freshwater, and saltwater. Featuring advanced inhibitor removal technology, the Quick-DNA/RNA™ Water Kit recovers high-quality DNA and RNA suitable for sensitive downstream applications such as NGS, qPCR, ddPCR, RT-qPCR, and RT-ddPCR.



The Quick-DNA/RNA™ Water Kit enables efficient purification of inhibitor-free nucleic acids from large water volumes. Designed for total nucleic acid extraction from viruses, bacteria, fungi, and other waterborne pathogens, it supports a wide range of sample types, including low-biomass water, raw wastewater, sewage, sludge, treated water, natural water, rivers, freshwater, and saltwater. Featuring advanced inhibitor removal technology, the Quick-DNA/RNA™ Water Kit recovers high-quality DNA and RNA suitable for sensitive downstream applications such as NGS, qPCR, ddPCR, RT-qPCR, and RT-ddPCR.
A Reliable, Streamlined Purification Solution
(I) Sample Preparation


Treat with Wastewater Stabilization Buffer
(II) Sample Lysis


Homogenize Using ZR BashingBead™ Lysis Tubes
(Recommended for complete microbial lysis)
(III) Sample Extraction


Utilizing Zymo-Spin™ technology
(IV) Inhibitor Removal


Featuring Zymo-Spin™ HRC Filters
Superior Viral Detection in Wastewater
A) SARS-CoV-2 RNA Detection by Digital PCR
A) Digital PCR quantified SARS-CoV-2 (N2 gene) in wastewater eluates extracted with the Quick-DNA/RNA™ Water Kit and other commercial kits (n=3) using the Bio-Rad™ PREvalence ddPCR™ SARS-CoV-2 Wastewater Quantification Kit.
B) RT-qPCR Assessment of Viral Recovery
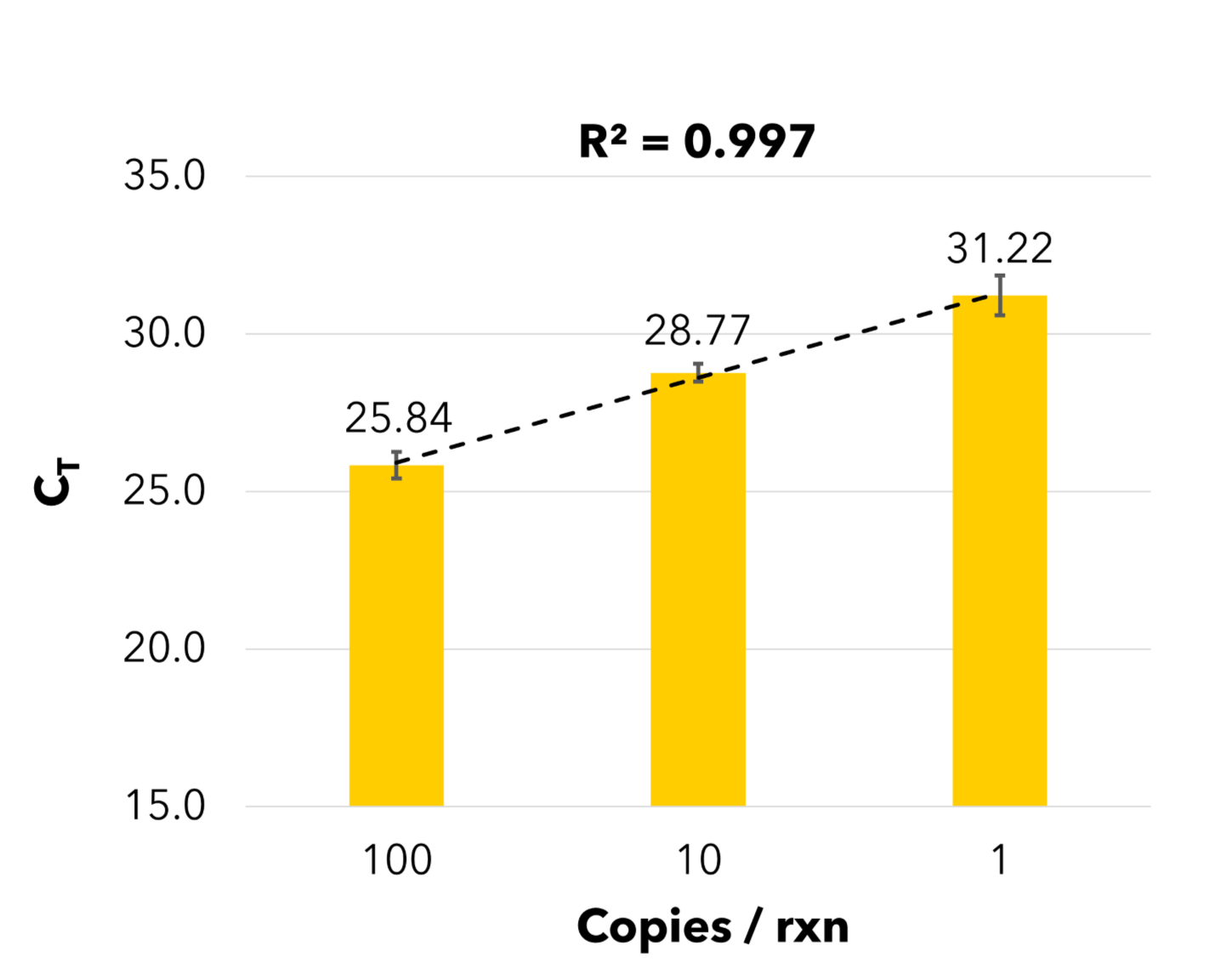
B) Viral RNA recovery was quantified by RT-qPCR using the Quick SARS-CoV-2 Multiplex Kit, shown as genome equivalent copies per PCR reaction. The Quick-DNA/RNA™ Water Kit is ideal for SARS-CoV-2 wastewater surveillance, enabling efficient viral nucleic acid recovery.
The Quick-DNA/RNA™ Water Kit enhances viral pathogen detection. A) Digital PCR quantified native SARS-CoV-2 (N2 gene) in wastewater eluates extracted with the Quick-DNA/RNA™ Water Kit and other commercial kits (n=3) using the Bio-Rad™ PREvalence ddPCR™ SARS-CoV-2 Wastewater Quantification Kit. B) Heat-inactivated SARS-CoV-2 was spiked into non-positive influent wastewater aliquots at varying concentrations. Viral RNA recovery was quantified by RT-qPCR using the Quick SARS-CoV-2 Multiplex Kit, shown as genome equivalent copies per PCR reaction. The Quick-DNA/RNA™ Water Kit is ideal for SARS-CoV-2 wastewater surveillance, enabling efficient viral nucleic acid recovery.
Sensitive Recovery of Bacterial & Fungal Nucleic Acid
The Quick-DNA/RNA™ Water Kit enables efficient bacterial and fungal DNA recovery from wastewater. Candida auris and Mycobacterium tuberculosis were spiked into 10 mL of influent wastewater aliquots at varying concentrations, with all targets absent in the native wastewater sample. Nucleic acids were then extracted and purified using the Quick-DNA/RNA™ Water Kit.
A) qPCR Quantification of Bacterial DNA
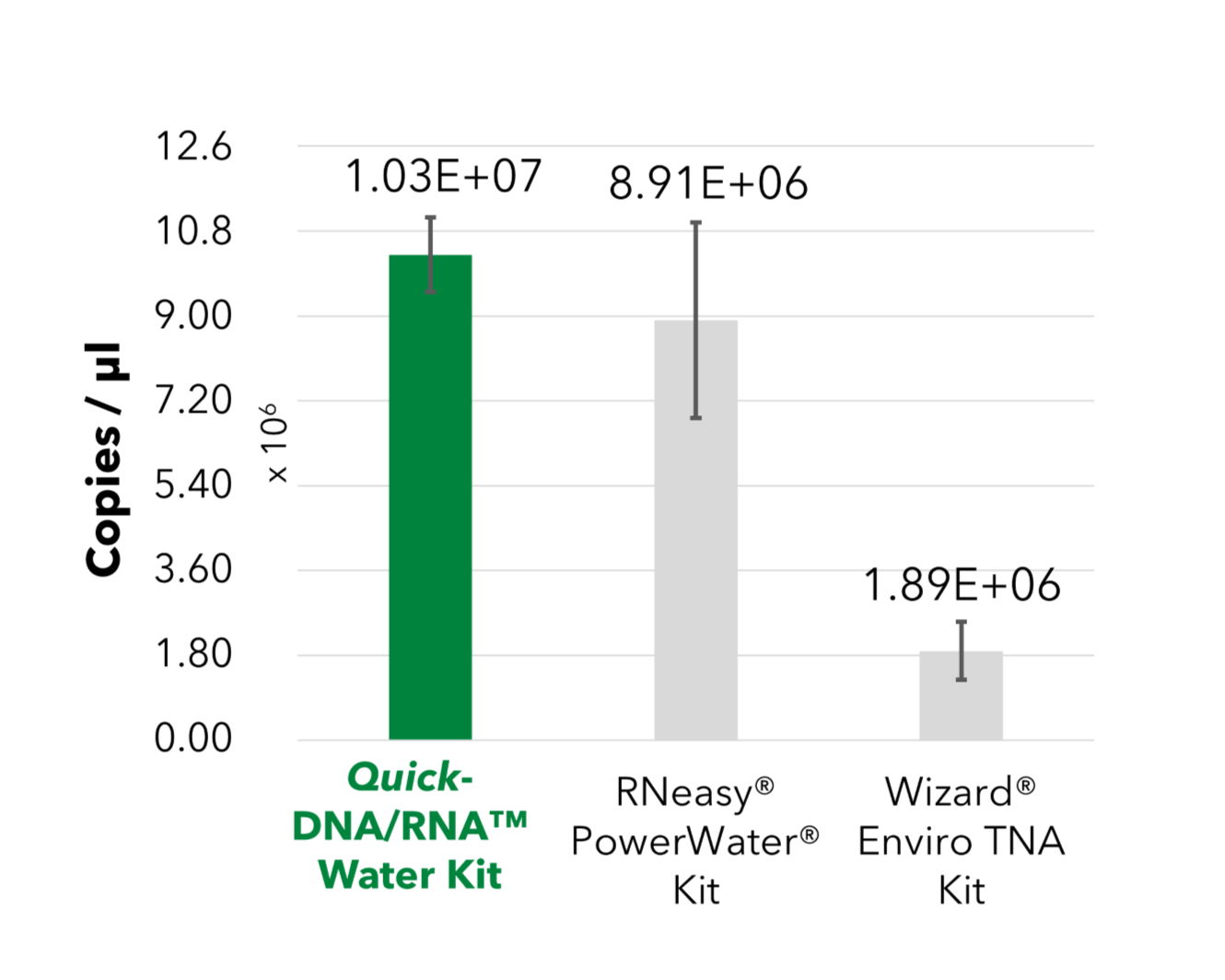
A) Bacterial DNA recovery was assessed and quantified by qPCR using the Femto Bacterial DNA Quantification Kit.
B) Fungi and Bacteria Detection using qPCR
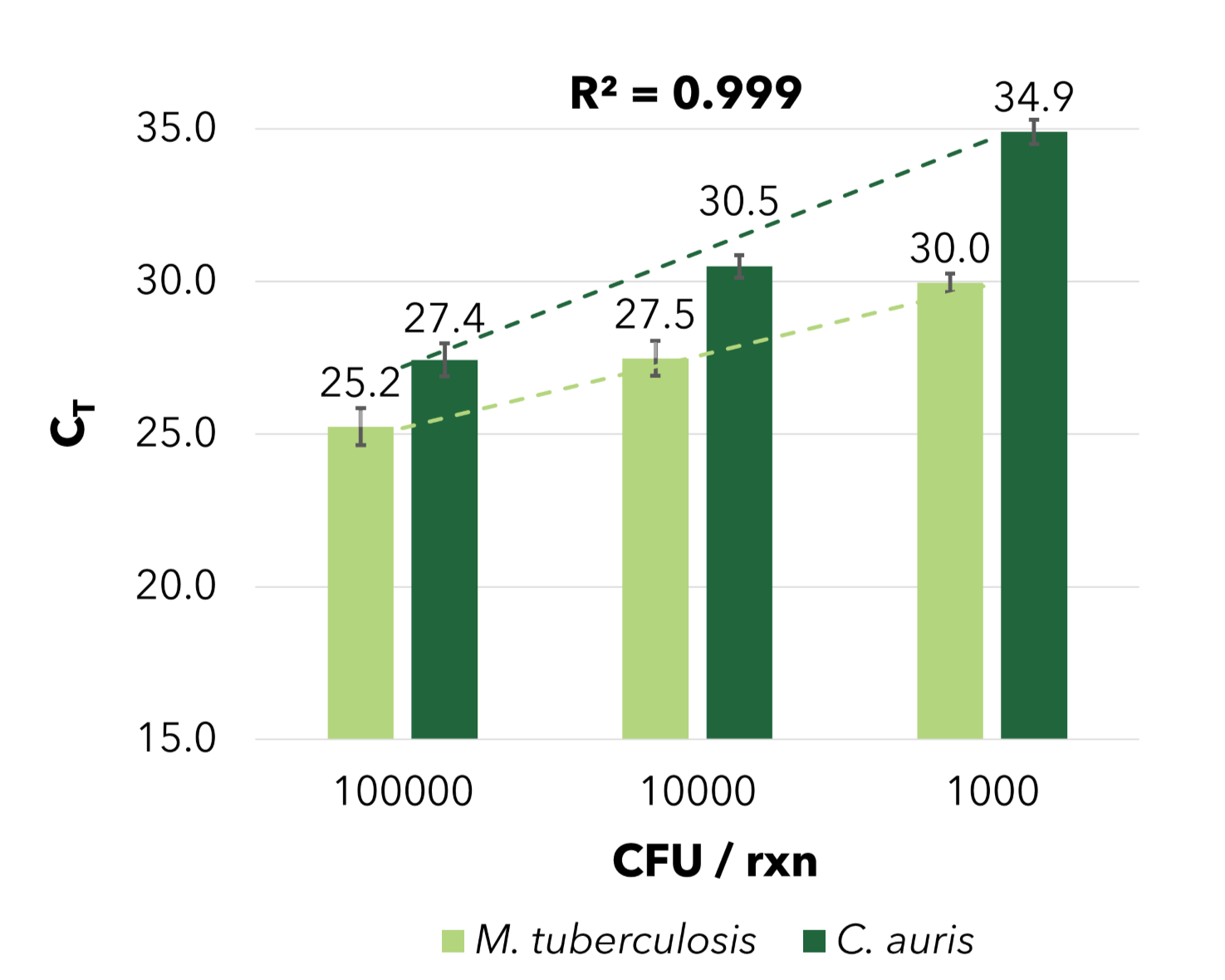
B) C. auris (fungi) and M. tuberculosis (bacteria) were measured by qPCR with target-specific primers, with results displayed as colony-forming units (CFU) per PCR reaction.
The Quick-DNA/RNA™ Water Kit enables efficient bacterial and fungal DNA recovery from wastewater. Candida auris and Mycobacterium tuberculosis were spiked into 10 mL of influent wastewater aliquots at varying concentrations, with all targets absent in the native wastewater sample. Nucleic acids were then extracted and purified using the Quick-DNA/RNA™ Water Kit. A) Bacterial DNA recovery was assessed and quantified by qPCR using the Femto Bacterial DNA Quantification Kit. B) C. auris (fungi) and M. tuberculosis (bacteria) were measured by qPCR with target-specific primers, with results displayed as colony-forming units (CFU) per PCR reaction.
Step 3: Molecular Data Generation
Microbiome Profiling
Our NGS library prep kits and services simplify microbiome profiling, minimizing hands-on time and streamlining wastewater surveillance workflows. Designed for efficiency, our ultra-fast library prep technology ensures a seamless process while delivering consistently accurate results. For a fully outsourced solution, our end-to-end NGS services cover every step, from DNA/RNA purification and library prep to sequencing and advanced bioinformatics analysis. Partner with our expert team to ensure precise, reliable data for your next wastewater-based epidemiology project.



Our NGS library prep kits and services simplify microbiome profiling, minimizing hands-on time and streamlining wastewater surveillance workflows. Designed for efficiency, our ultra-fast library prep technology ensures a seamless process while delivering consistently accurate results. For a fully outsourced solution, our end-to-end NGS services cover every step, from DNA/RNA purification and library prep to sequencing and advanced bioinformatics analysis. Partner with our expert team to ensure precise, reliable data for your next wastewater-based epidemiology project.
The Simplest and Fastest NGS Library Prep Procedures
16S/ITS Amplicon Sequencing
The Quick-16S Plus and Quick-ITS Plus NGS Library Prep Kits support amplification of the V1-V2, V1-V3, V3-V4, V4, and ITS2 regions. These kits feature a simplified, normalization-free protocol that requires only a single PCR step, enabling library preparation to be completed in just 30 minutes for up to 96 samples.
Whole Genome Sequencing
The Zymo-Seq SPLAT DNA Library Kit ensures true end ligation for reliable and precise genomic data across diverse research applications. The two-step workflow allows for the preparation of DNA samples into sequencing-ready libraries in as little as 3 hours, significantly speeding up the research process.
Metatranscriptomic Sequencing
The Zymo-Seq RiboFree® Total RNA Library Kit features a unique universal depletion technology to efficiently remove rRNA from total RNA of any organism, enabling comprehensive gene expression analysis. Its automation-friendly protocol streamlines the process, allowing RNA to be converted into NGS libraries in as little as 4 hours.
End-to-End Sequencing Services for the Most Accurate Microbiome Profiling
Shotgun Metagenomic Sequencing
Gain unparalleled strain-level resolution with our Shotgun Metagenomic Sequencing Service. Our comprehensive deliverables include taxonomy analysis, alpha diversity bar plots, beta diversity analysis, gene family profiling, metabolic pathway insights, antibiotic resistance and virulence gene profiling, and LEfSe biomarker discovery.
Whole Genome Sequencing
Unlock the full genetic blueprint of microbial pathogens with our Whole Genome Sequencing (WGS) Service. Ideal for public health, outbreak investigations, and surveillance, our WGS platform enables high-resolution tracking of transmission, antimicrobial resistance, and emerging variants.
16S/ITS Amplicon Sequencing
Obtain precise species-level identification and absolute abundance quantification with our 16S/ITS Amplicon Sequencing Service. Our streamlined workflow ensures industry-leading turnaround times, delivering high-quality sequencing results in less than a week.
Full-Length 16S Sequencing
Achieve unmatched species-level taxonomic resolution with PacBio HiFi sequencing and advanced bioinformatics analysis. Our Full-Length 16S Sequencing Service enables precise differentiation of closely related species, delivering high-quality results in as little as 3 weeks.
Metatranscriptomic Sequencing
Uncover intricate microbial dynamics with our Metatranscriptomic Sequencing Service, which provides comprehensive metabolic pathway and gene expression analysis. Utilizing our proprietary universal rRNA depletion technology, RNA can be detected and sequenced across all kingdoms: bacteria, archaea, eukarya, and viruses.
Long-Read Metagenome Sequencing & Assembly
Our Long-Read Metagenome Sequencing & Assembly Service includes sequence assembly, binning, taxonomy assignment, and composition profiling. Combining our unbiased lysis protocol, PacBio HiFi sequencing, and bioinformatics, this service consistently delivers accurate profiling.
Targeted Panels & Assays



High Sensitivity Microbial and Pathogen Detection from Wastewater
Step 4: Bioinformatic Analysis
Pathogen Monitoring



Zymo Research's bioinformatics pipeline for wastewater surveillance enables comprehensive detection and quantification of microbial pathogens, including viruses, bacteria, and antimicrobial resistance genes. These tools support high-resolution tracking of variants and strains, offering critical insights into pathogen evolution and transmission. By integrating sequencing data with temporal and spatial metadata, they also uncover trends in microbial community dynamics, providing valuable information for public health decision-making and outbreak response.
VirSieve
Open-source bioinformatics pipeline ideal for detecting viral variants.
VirSieve is an open-source bioinformatics tool designed for high-sensitivity genomic variant detection in challenging sample matrices, such as wastewater. While currently optimized for SARS-CoV-2, the tool is adaptable to a wide range of viral species, providing a streamlined approach to detecting and identifying viral mutations. The pipeline automates key steps, including alignment, primer trimming (if needed), variant calling, and annotation, with a strong emphasis on distinguishing true biological mutations from sequencing artifacts or spurious results. Built around software containers, VirSieve includes pre-built reference data to ensure consistency across runs and between laboratories.



VirSieve is an open-source bioinformatics tool designed for high-sensitivity genomic variant detection in challenging sample matrices, such as wastewater. While currently optimized for SARS-CoV-2, the tool is adaptable to a wide range of viral species, providing a streamlined approach to detecting and identifying viral mutations. The pipeline automates key steps, including alignment, primer trimming (if needed), variant calling, and annotation, with a strong emphasis on distinguishing true biological mutations from sequencing artifacts or spurious results. Built around software containers, VirSieve includes pre-built reference data to ensure consistency across runs and between laboratories.
Wastewater Bioinformatic Pipeline Comparison
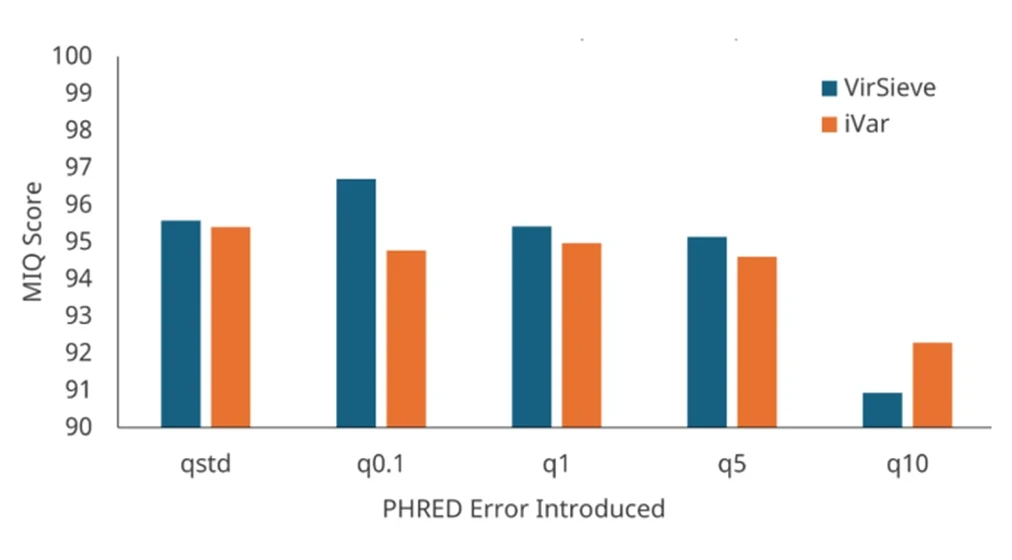
Comparison of VirSieve and iVar bioinformatics pipelines for SARS-CoV-2 wastewater surveillance and strain identification under increasing PHRED error levels. VirSieve achieves higher MIQ Scores than iVar, particularly at low error rates (q0.1), demonstrating greater reliability in handling sequencing noise.
Tell Us More About Your Wastewater Project

Tell Us More About Your Wastewater Project
In line with our 100% Customer Satisfaction Guarantee, our team of scientists is ready to provide tailored guidance and dedicated support for your wastewater surveillance project. Schedule a consultation today to learn how partnering with Zymo Research can elevate your research while saving you time and expense.
Connect with a ScientistTrusted by Leading Public Health Groups
We are proud to have served institutions across the United States to support wastewater surveillance workflows that keep our communities safe.




Featured Publications & Citations
Wastewater Surveillance Resources
A Modern Approach to Pathogen and AMR Detection
Learn about the role of wastewater surveillance in public health and explore solutions that ensure accuracy and reliability across every stage of the workflow.
Insights from the Microbes in Wastewater Symposium 2025
Review the key themes from the Microbes in Wastewater Symposium 2025, from pressing challenges like climate change and lack of standardization to emerging opportunities in microbiome research and bioinformatics.
An Integrated Approach for Wastewater Surveillance
Discover how the Quick-DNA/RNA™ Water Kit combined with short-read sequencing platforms enhances pathogen detection, antimicrobial resistance (AMR) monitoring, and functional analysis in wastewater systems.
Sequencing RNA from Southern California Wastewater
Hear from Dr. Jason Rothman, Assistant Professor of Teaching at the University of California, Riverside, on how sequencing wastewater nucleic acids provides valuable insights into microbial ecology and aids in detecting potential pathogens.
Nucleic Acid Extraction of Pathogens in Wastewater
Learn how combining Ceres Nanosciences’ Nanotrap® Microbiome Particles and Enhancement Reagents with Zymo Research’s Quick-DNA/RNA™ Water Kit creates a powerful, integrated workflow for enhanced wastewater monitoring.
What Are PCR Inhibitors?
Learn how to remove organic and inorganic contaminants from environmental samples like wastewater to prevent interference in PCR and NGS reactions, ensuring the purification of high-quality DNA and RNA for accurate results.
An Integrated Approach for Wastewater Surveillance
Discover how the Quick-DNA/RNA™ Water Kit combined with PacBio Onso short-read sequencing enhances pathogen detection, antimicrobial resistance (AMR) monitoring, and functional analysis in wastewater systems.
Understanding Raw NGS Data
Hear from Dr. Michael Weinstein, Director of Systems Biology at Zymo Research and Adjunct Faculty Member at the University of California, Los Angeles, as he breaks down the fundamentals of handling NGS output files.
Exploring Microbiome Profiling Techniques
Compare and contrast 16S rRNA Gene Amplicon Sequencing and Shotgun Metagenomic Sequencing, focusing on their differences in microbiome profiling depth and breadth as well as the accessibility of each workflow.
Microbiome Assessment of Water and Sewage Treatment Plants
Explore the unbiased microbial profiling workflow that enables accurate and reliable NGS-based analysis of environmental water, tap water, and wastewater from sewage treatment plant influent and effluent.
Innovators & Pioneers
Leveraging Wastewater & NGS for Early Virus Detection
Discover how researchers combined PCR inhibitor removal, next-generation sequencing (NGS), and an innovative bioinformatics approach to track emerging SARS-CoV-2 strains in wastewater samples from multiple treatment plants in Switzerland.
A Clear Future for Dirty Water
Learn how the Advanced Water Purification Facility (AWPF) in Orange County, California utilizes NGS-based monitoring to verify the safety of recycled wastewater and mitigate public health risks.
Assessing Fecal Pollution in Urban Watersheds
Read about how researchers from the US Environmental Protection Agency (EPA) and the University of Alabama investigated fecal contamination in Turkey Creek, aiming to better understand the sources and dynamics of this pollution.
How Yale Monitors COVID-19 Surges in Wastewater
Discover how Yale’s Environmental Engineering Program launched a CDC-funded project to monitor SARS-CoV-2 levels in Connecticut’s wastewater, using robust RNA extraction technology to detect and track COVID-19 outbreaks.
LA Wastewater Tells A Story of Monitoring COVID-19
Learn how the Los Angeles County Sanitation District (LACSD) monitors SARS-CoV-2 levels in wastewater, using reliable sample collection and RNA extraction reagents to identify communities at risk of widespread infection.

