RNA sequencing (RNA-Seq) utilizes next-generation sequencing technology to determine the quantity and nucleotide sequences of expressed transcripts within a sample or organism. With its capability to produce massive amounts of data, RNA-Seq has become an indispensable tool and widely adopted in many research fields. A variety of RNA-Seq methods have been developed over the years, and it is critical to choose the most suitable method as it directly impacts the data that researchers acquire. Herein, we describe the features of two common RNA-Seq methods, total RNA-Seq and 3’ mRNA-Seq, to assist in the selection of the RNA-Seq method that can better fulfill your research needs.
The features of both methods largely come from the difference in their respective library preparation strategies. RNA-Seq library preparation starts with RNA extracted from sample sources such as tissue or cell cultures. In eukaryotes, the extracted RNA consists of many RNA types including mRNAs which encode proteins and are mostly polyadenylated, and non-coding RNAs such as rRNA, lncRNAs, tRNA, miRNA, and snoRNA. The rRNA is massively overrepresented, comprising 80-90% of the molecules found in total RNA samples.1
-
Total RNA-Seq usually depletes rRNA from the input RNA and leverages random primers to produce cDNA from both coding and non-coding RNAs. The generated cDNAs cover the transcripts quite evenly, without obvious bias towards either 5’ or 3’ ends (Figure 1A). An example total RNA-Seq library prep method is found in the Zymo-Seq RiboFree® Total RNA Library Kit.
-
3’ mRNA-Seq typically uses oligo-dT primers to target mRNA via its poly-A tail directly. Paired with other steps in the workflow, the produced cDNA is biased towards the 3’ end of transcripts (Figure 1B). An example 3’ mRNA library prep method is found in the Zymo-Seq SwitchFree™ 3’ mRNA Library Kit.
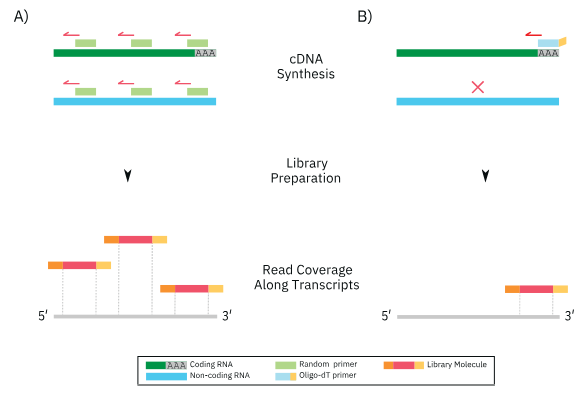
As a result, these two-methods profile different ranges of RNA types and cover different regions across the transcripts, making them ideal for different applications.
Total RNA-Seq quantifies both coding and non-coding transcripts, with information obtained across the entire length of transcripts. Hence, it can be used to examine global transcript expression, splicing patterns, exon/intron boundaries, and RNA regulation.2 This broad coverage provides great versatility and is useful to those interested in examining whole-transcriptome level changes and interactions.
3’ mRNA-Seq delivers a narrower scope of information by focusing on protein coding genes. It is mainly useful for differential gene expression (DGE) analysis, one of the most common RNA-Seq applications3 that quantifies gene expression changes between different biological conditions.
Additionally, the distinct library prep principles of total RNA-Seq and 3’ mRNA-Seq determine their difference in tolerating degraded RNA input such as those extracted from FFPE tissues: 3’ mRNA-Seq may not capture protein coding transcripts as effectively as total RNA-Seq due to the likely lack of intact poly-A tails in the RNA extracted from these samples.
Furthermore, the minimum sequencing depth requirement is also different between the two methods. A study showed that total RNA-Seq required ~ 3X the number of reads per library compared to mRNA-Seq for the same transcriptome coverage.4 This is mainly due to total RNA-Seq’s broad capability in detecting both coding and non-coding RNA. The generally lower sequencing depth requirements of mRNA-Seq methods allow for multiple 3’ mRNA-Seq libraries to be sequenced for the same price of one total RNA-Seq library. However, 3’ mRNA-Seq libraries can be sequenced at higher depths if desired. In addition to the method, the optimal sequencing depth depends on other factors such as sample species, quality of input, experimental goals, and budget. It is recommended to consult the literature and your sequencing provider for suggestions on sequencing depth.
Taken together, evaluating your specific research goal is a top consideration when selecting between total RNA-Seq and 3’ mRNA-Seq. Each covers different RNA types and transcript regions, thus making them suitable for different applications. The input RNA’s integrity and the resources at one’s disposal also play an important role in choosing an applicable RNA-Seq method for a well-designed project.
| Total RNA-Seq | 3’ mRNA-Seq | |
|---|---|---|
| RNA Type of Interest |
|
|
| Transcript Coverage |
|
|
| Suggested Applications |
|
|
| Recommended Zymo-Seq Library Kit | Zymo-Seq RiboFree® Total RNA Library Kit | Zymo-Seq SwitchFree™ 3’ mRNA Library Kit |
Citations
- O'Neil, D.; Glowatz, H.; Schlumpberger, M. Ribosomal RNA depletion for efficient use of RNA-seq capacity. Curr Protoc Mol Biol 2013, Chapter 4, Unit 4.19. DOI: 10.1002/0471142727.mb0419s103.
- Hrdlickova, R.; Toloue, M.; Tian, B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip Rev RNA 2017, 8 (1). DOI: 10.1002/wrna.1364.
- McDermaid, A.; Monier, B.; Zhao, J.; Liu, B.; Ma, Q. Interpretation of differential gene expression results of RNA-seq data: review and integration. Brief Bioinform 2019, 20 (6), 2044-2054. DOI: 10.1093/bib/bby067.
- Zhao, W.; He, X.; Hoadley, K. A.; Parker, J. S.; Hayes, D. N.; Perou, C. M. Comparison of RNA-Seq by poly (A) capture, ribosomal RNA depletion, and DNA microarray for expression profiling. BMC Genomics 2014, 15 (1), 419. DOI: 10.1186/1471-2164-15-419.
