What is PCR?
Polymerase chain reaction (PCR) is an essential molecular biology technique used to amplify specific segments of DNA. PCR was first developed by Kary Mullis who won the Nobel Prize for Chemistry in 1993 for his revolutionary method.1 PCR is essential for a wide range of applications such as genetic mapping, bacterial & viral detection, cloning, sequencing, and forensics.
Although there is a vast array of specialized PCR methods, the general workflow and components for the reaction remain the same. PCR relies on fluctuations in temperature achieved by a thermal cycler and Taq polymerase, a special polymerase that is functional even at elevated temperatures. The necessary components for any PCR reaction are a DNA template strand, oligonucleotide primers, deoxynucleotide triphosphates (dNTPS), buffer, and Taq polymerase.2 A general PCR amplification cycle follows the same principal steps:
- Denature DNA – The template DNA is heated above 94 °C to separate the double stranded DNA (Figure 1).
- Anneal Primers – The temperature is then lowered to between 40-60 °C, enabling the primers to anneal to the separated DNA strands at a specific sequence (Figure 1).
- Extend Primers – Lastly, the temperature is increased to an optimal 70-74 °C, enabling Taq polymerase to elongate the new strand from the primer by inserting nucleotides complementary to the template DNA in the 5’ to 3’ direction (Figure 1).
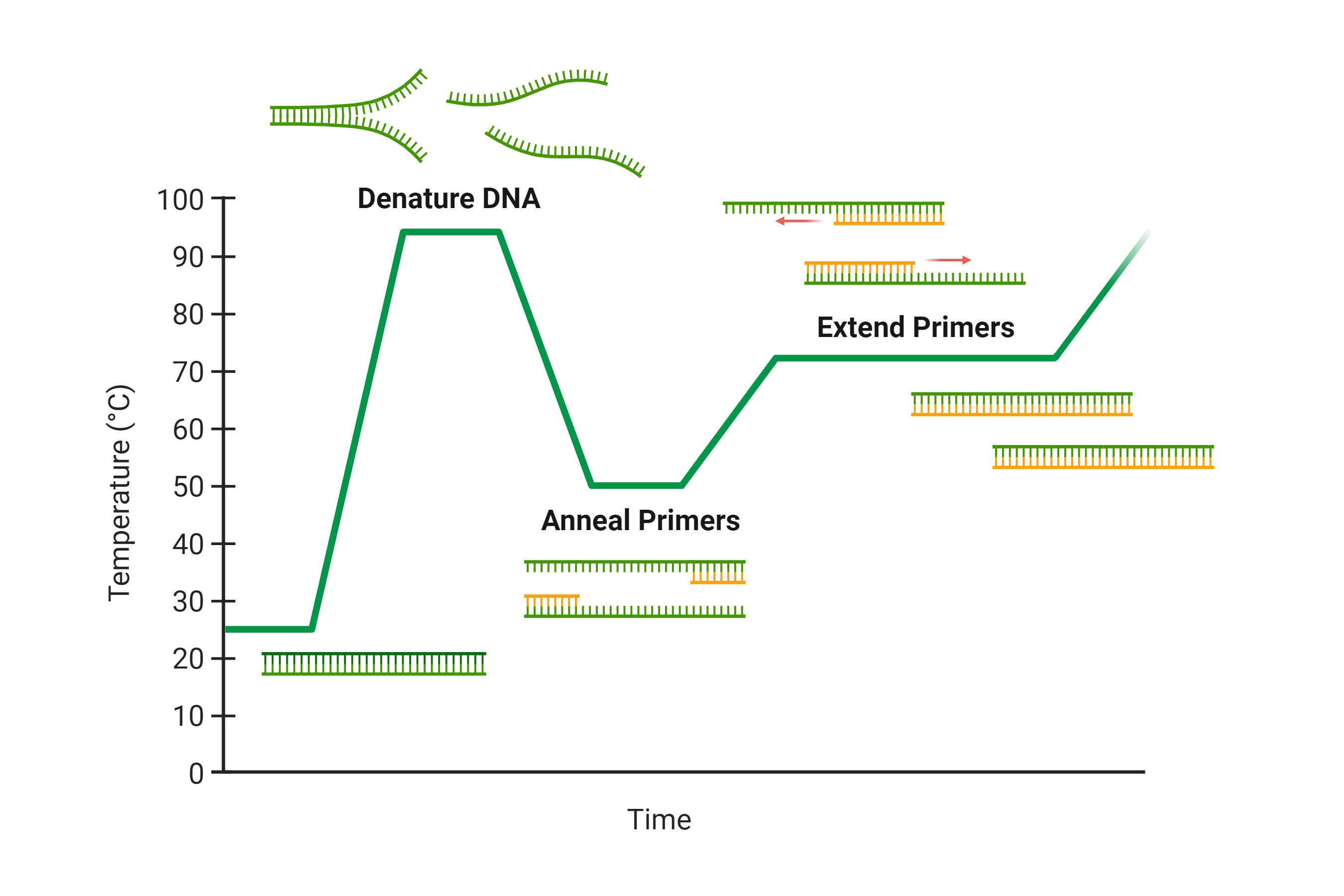
Figure 1. Overview of the key steps during PCR and the corresponding temperatures they take place at.
The PCR cycle will repeat anywhere from 20-40 times to yield millions of copies of the desired DNA strand. Highly specific primers are critical during the annealing step to obtain sufficient yields of your desired PCR product and to avoid amplification of off-target sequences. Read our complete guide on How to Design Primers for more information on how to design effective PCR primers for different kinds of PCR.
Real Time PCR (qPCR)
When PCR was first developed, PCR products could only be quantified after the reaction was complete by using endpoint analysis techniques, like gel electrophoresis. However, an alternate method known as quantitative Real Time PCR (or simply qPCR) was developed to quantify your PCR product as the reaction is running. During qPCR, fluorescent reporter molecules (e.g. DNA binding dyes, fluorescently labeled PCR primers or probes) are incorporated into the DNA strand to quantify the PCR product in real time using fluorescence.3 The resulting qPCR curve can be used to monitor amplification in real time (Figure 2). Special thermal cyclers can detect fluorescence and perform amplification simultaneously to accurately detect and quantify fluorescence with each amplification cycle. The benefits of qPCR over end-point PCR include the ability to accurately quantify initial template DNA input as well as increased sensitivity. To save time, researchers commonly use qPCR premix kits that provide many of the reaction components in just one tube, which streamlines the preparation of the reaction.
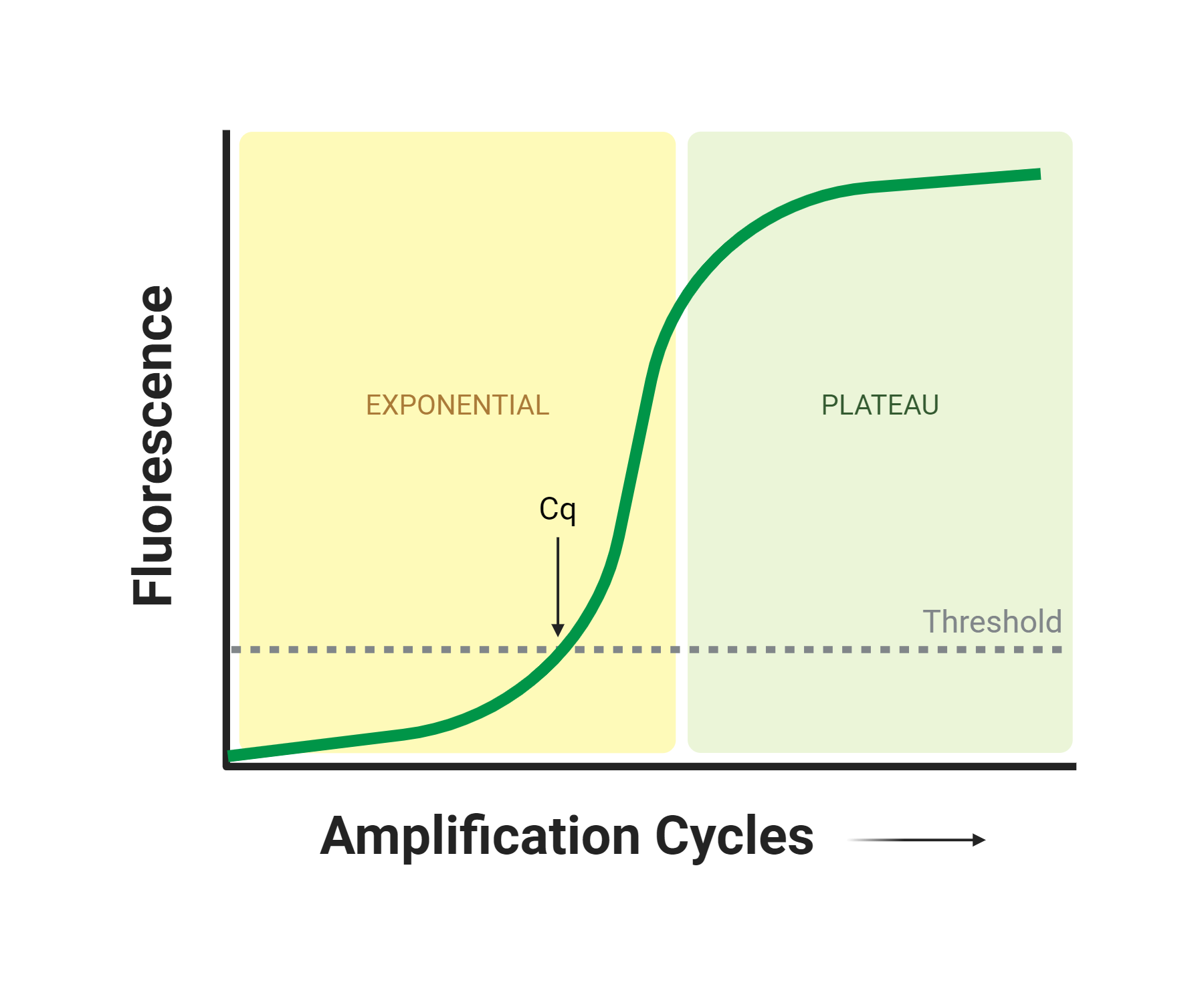
Figure 2. An example of a qPCR Curve. As an increasing number of PCR cycles are performed, the amount of PCR product increases exponentially, resulting in increasing fluorescence until the fluorescence eventually plateaus as the reaction reagents are depleted. The Cq (quantification cycle) is the number of cycles at which the fluorescence level is above a pre-defined threshold, set to determine amplification from background fluorescence. The Cq value can be used to quantify the starting DNA input and final amplicon output if standards are run in parallel to generate a standard curve.
RT-PCR
For several applications, such as RNA virus detection or gene expression studies, it is necessary to amplify RNA segments instead of DNA. RNA can be amplified using PCR, but it first must be converted to cDNA to create a suitable template for Taq polymerase. A reverse transcriptase is employed to convert RNA to cDNA, hence the name RT-PCR (reverse transcription PCR). There are two approaches to RT-PCR: one-step and two-step RT-PCR. During one-step RT-PCR, the reverse transcription and PCR amplification step occur in the same tube while during two-step RT-PCR the reverse transcription and amplification steps are done separately. One-step RT-PCR is more efficient and well suited for high-throughput sample processing. However, if the PCR parameters need to be fine-tuned, two-step RT-PCR is the preferred method. Choosing the proper RT-PCR method will depend on your desired throughput and application. There are commercially available RT premix kits that include all the necessary reagents in a ready-to-use master mix for simplifying the cDNA synthesis step.
Droplet Digital PCR (ddPCR)
Droplet digital PCR is a variation of PCR that utilizes a water-oil emersion to separate amplified PCR products. The partitioning of accumulating amplicons works like wells on a 96-well plate but instead of 96-wells the sample is separated into 20,000 droplets, enabling the precise quantification of DNA molecules without a reference standard.4 The molecule-level sample processing enables decreased sample input requirements compared to other PCR methods and incredibly sensitive detection of slight differences in amplicon count between samples.
Hot Start PCR
At room temperature, PCR primers can anneal to regions of the template DNA that are not perfectly complementary. If Taq polymerase elongates these off-target primers, undesired PCR products will be generated. To mitigate amplification of off-target DNA segments, researchers can employ a “hot start” DNA polymerase that has been impaired with chemical modifiers to block the polymerase’s enzymatic activity at room temperature. When thermal cycling begins and the temperature reaches 94 °C the covalent bonds adhering the modifier to the DNA polymerase break, restoring the functionality of the polymerase. It is also possible to perform hot start PCR manually by leaving out a component of the PCR reaction until the reaction is heated to 60 °C.5
Multiplex PCR
Multiplex PCR makes it possible to amplify multiple targets at once by employing two or more pairs of PCR primers. Multiplex PCR is an efficient technique that enables the comparison of multiple amplicons simultaneously. However, optimizing annealing conditions and ensuring high primer specificity is especially important for successful multiplex PCR. Some of the challenges to overcome when doing multiplex PCR include non-specific amplification and reduced efficiency if there is any competition between the primers. If considering doing a multiplex reaction, ensure your primers are highly unique and that your amplicons are different sizes so they can be distinguished on a gel.
Nested PCR
Nested PCR uses two pairs of PCR primers for improved yield and specificity of amplicons, making this technique effective for amplifying low-input samples or samples that require increased specificity.6 During the first round of PCR, the DNA strand is denatured and then the first pair of primers anneal to outer primer sites that flank the amplicon of interest (Figure 3). The product of this initial round of PCR is then amplified again using a second pair of primers that bind to primer sites that are “nested” within the beginning and end of the DNA sequence of interest, hence the term nested PCR (Figure 3.). The two rounds of amplification halt the production of non-specific products because in the off chance an off-target segment is amplified in the first round of PCR, it is very unlikely that the non-specific region will be recognized and amplified by the nested primer set during the second round of PCR.
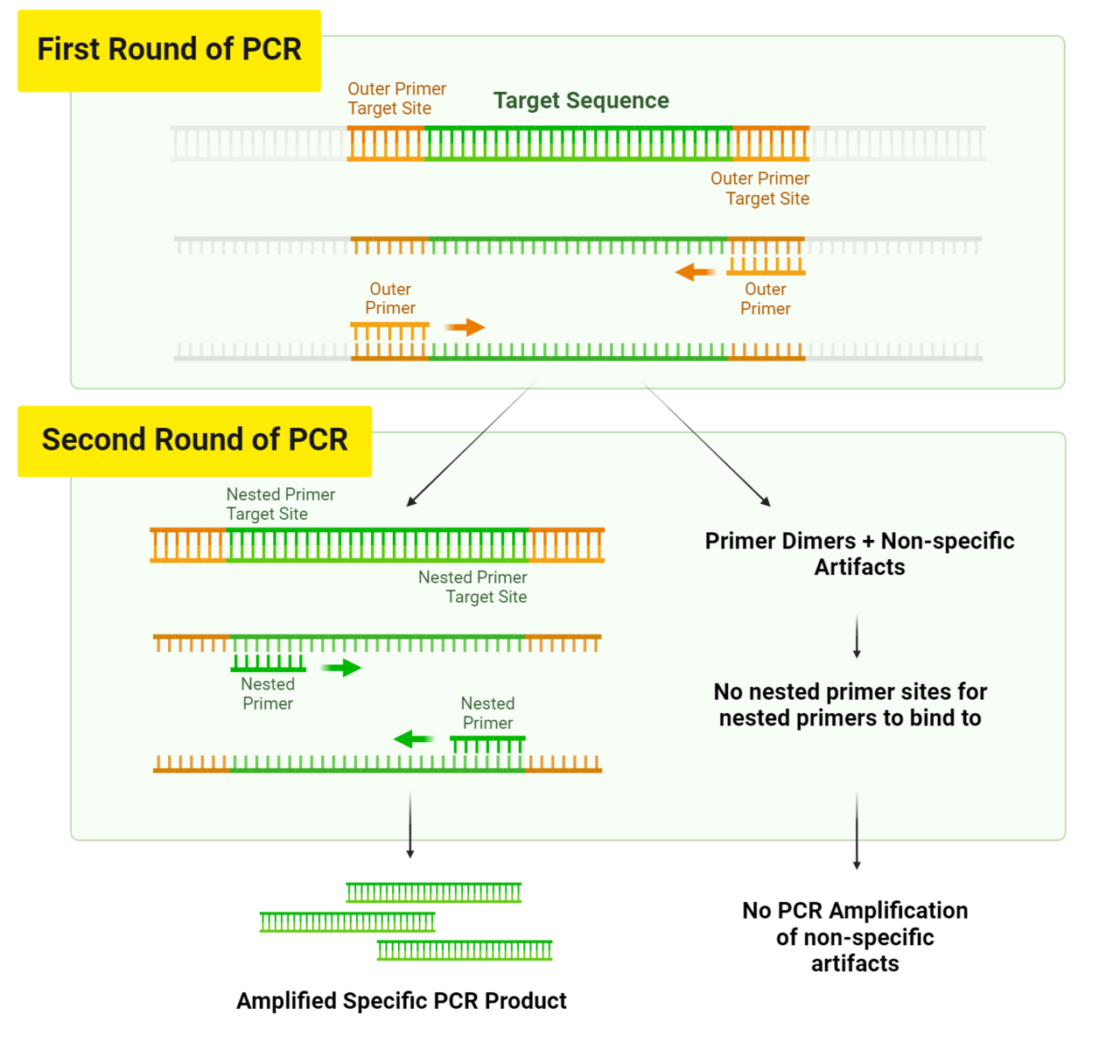
Figure 3. How Nested PCR is Used to Overcome Low Specificity. Nested PCR ensures that there are no primer dimers or non-specific products after the second round of amplification, as any non-specific PCR products from the first round of amplification will not have the nested primer sites necessary to be amplified during the second round of PCR.
Your PCR Reaction is Only as Good as Your Template
PCR amplification is a fundamental molecular biology tool that is essential to many downstream applications such as cloning and NGS. However, no matter the application, the best way to set yourself up for PCR success is to start with high-quality template nucleic acid. Zymo Research’s Quick-DNA and Quick-RNA kits are robust extraction solutions that ensure eluted nucleic acid is high yield, ultra-pure and suitable for PCR amplification. In addition, if you are working with samples high in polyphenolics, like plants, soil, or skin tissue, the OneStep PCR Inhibitor Removal Kits are ideal for the complete removal of inhibitors for highly efficient PCR amplification. If you aren’t getting the purity or concentration you need from your existing extraction method, the DNA Clean & Concentrator Kits can be used to clean-up template DNA to be used for PCR or to completely remove salts, polymerases, and endonucleases from your PCR product, leaving you with DNA that is suitable for even the most sensitive downstream applications. If your workflow involves extracting a PCR product from agarose gel, the Zymoclean Gel DNA Recovery Kits ensure your recovered DNA is suitable for highly sensitive downstream applications including sequencing, ligation, and endonuclease digestion. Finally, the ZymoPURE Plasmid Purification Kits are ideal for producing plasmid that is pure enough for PCR reactions or transfections. Discover which DNA and RNA purification kit best suits your research goals today.
Citations
- National Human Genome Research Institute. (2020). Polymerase Chain Reaction (PCR) fact sheet. Retrieved December 5, 2024, from https://www.genome.gov/about-genomics/fact-sheets/Polymerase-Chain-Reaction-Fact-Sheet
- Lorenz T. C. (2012). Polymerase chain reaction: basic protocol plus troubleshooting and optimization strategies [Video]. Journal of Visualized Experiments.https://doi.org/10.3791/3998
- Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., Mueller, R., Nolan, T., Pfaffl, M. W., Shipley, G. L., Vandesompele, J., & Wittwer, C. T. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical chemistry, 55(4), 611–622. https://doi.org/10.1373/clinchem.2008.112797
- Quan, P. L., Sauzade, M., & Brouzes, E. (2018). dPCR: A Technology Review. Sensors, 18(4), 1271. https://doi.org/10.3390/s18041271
- Green, M. R., & Sambrook, J. (2018). Hot Start Polymerase Chain Reaction (PCR). Cold Spring Harbor protocols, 2018(5), 10.1101/pdb.prot095125. https://doi.org/10.1101/pdb.prot095125
- Green, M. R., & Sambrook, J. (2019). Nested Polymerase Chain Reaction (PCR). Cold Spring Harbor protocols, 2019(2), 10.1101/pdb.prot095182. https://doi.org/10.1101/pdb.prot095182
