Key Takeaways
-
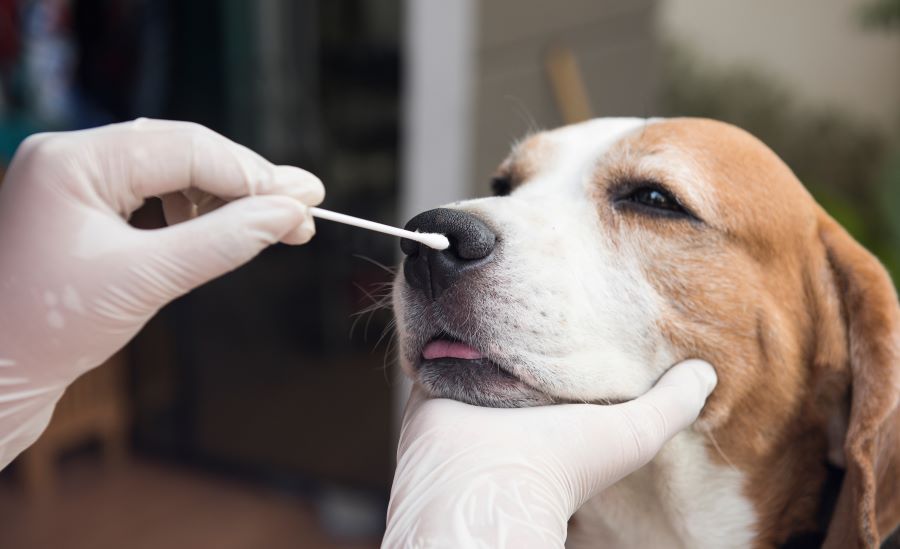
MiDOG Animal Diagnostics Company uses NGS-based diagnostic testing to provide accurate and high-throughput pathogen detection for all animals. Here’s a sneak peak of how they use Zymo Research products to enhance their workflow.
-
Challenges Solved:
- Accurate pathogen identification (bacteria, fungi, AMR, etc.)
- Fast turnaround time (48 hrs.)
- Standardized and quality-controlled workflow
-
Microbiome Solutions Mentioned:
- Sample Collection: DNA/RNA Shield Stabilization Reagent
- DNA/RNA Extraction: ZymoBIOMICS DNA/RNA Kits
- Microbiome Standards & Mock Communities: ZymoBIOMICS Standards
- NGS Library Prep: Quick-16S & ITS NGS Kits
Value of NGS-Based Diagnostic Testing
When it comes to accurate pathogen identification, NGS-based testing has many advantages over traditional culture and PCR testing. The application is rapidly being adopted by general veterinary practitioners and specialty clinics such as dermatology, urology, eye clinics, exotic pet clinics, zoos, and aquariums.
Some of the main advantages of NGS-based testing include:
- Accurate pathogen identification based on the genomic profile rather than cytological appearance
- Simultaneous detection of complex multi-organism pathogenic mixtures like biofilms
- Novel pathogen diagnosis
- Antibiotic resistance prediction, particularly of unculturable pathogens
- Ability to screen for any number of pathogens without limitations
MiDOG Animal Diagnostics Company has been providing this testing method to veterinarians across North America for the last 5 years. Here’s a sneak peak of how they use Zymo Research’s microbiome products to enhance their workflow.
A Standardized, Quality-Controlled Workflow
Standard
Collection
Extraction
Preparation

From the time the veterinarian collects a sample, the main goals of the MiDOG testing workflow are:
- To ensure accuracy of the microbial profile being reported
- Specificity of the pathogens detected
- A quick turnaround time (48 hrs.)
To do this effectively, a few key factors need to be addressed at each step of the workflow to prevent bias or loss of information.
-
Sample Collection
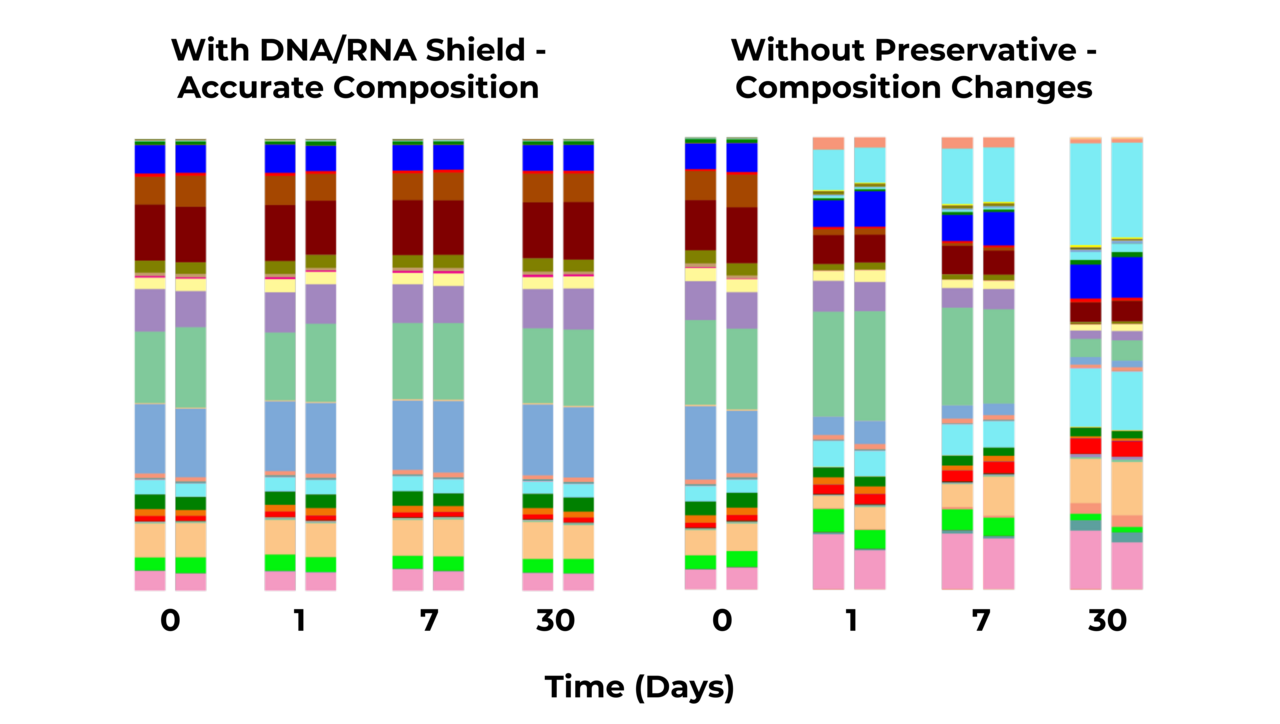
Figure 1. Microbial composition of stool is unchanged after one month at ambient temperature with DNA/RNA Shield. The extracted DNA was subjected to microbial composition profiling via 16S rRNA gene targeted sequencing.
Everything starts with a sample from a pet that is most often suffering from an infection. These infections can include wounds, otitis, dermatitis, diarrhea, urinary tract infections and more. The variety of sample types an NGS-based diagnostic lab may receive knows almost no limits! Having a collection method that can accommodate tissue samples from feline wounds, swabs of dolphin blow wholes, and feces from a rhino is essential.
What all these samples have in common is that the microbial profile needs to be preserved just as it was at the time the clinician collected the sample. It can get hectic in the clinic and a sample may sit idle for a while, which can introduce bias to the microbial profile (Figure 1). A solution that can protect the samples at room temperature and can handle any kind of sample type is the DNA/RNA Shield Stabilization Reagent. DNA/RNA Shield is very easy to use, as the clinician or technician simply adds the swab, feces, nasal wash, tissue, etc. to the buffer and it is stable and ready to go.
Benefits of DNA/RNA Shield Stabilization Reagent include:
-
The microbial profile remains the exact same as it was when it was collected from the patient, no growth or decay of certain pathogens or commensals can occur
-
Temperature resistant: no matter if the sample is collected in a mobile pet clinic in the heat of a Texas summer or by a research group collecting loon fecal samples in Alaska, no special precaution or refrigeration is needed
-
Samples are stable for up to 30 days at room temperature, or up to 2 years when frozen. So, no worries if a sample has accidentally been forgotten over the weekend.
-
Pathogen inactivation, allowing for safe handling of the sample by clinicians, technicians, and the shipping service provider.
After the sample has been safely transported to the lab, the next steps of the processing can begin.
-
-
DNA Extraction
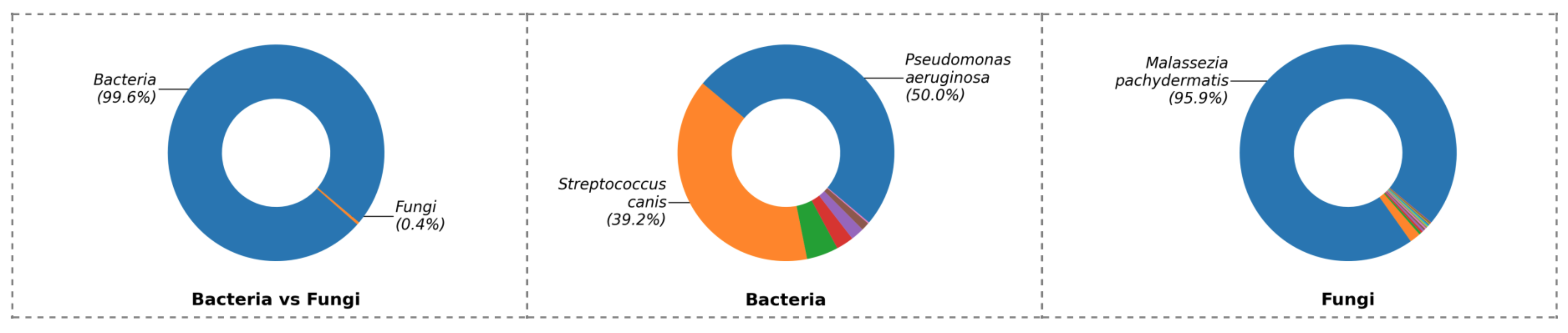
Figure 2. Example MiDOG report. In each box - Bacteria vs. Fungi: the relative abundance between Bacteria and Fungi. Bacteria: the percentage profile of bacterial species alone. Fungi: the percentage profile of fungi species alone. Each color represents a species. The larger the colored segment is, the more abundant the species is.
To accurately report all the pathogens and commensals present in the patient samples, the DNA (and/or RNA) from all those microbes must be extracted. It’s not simply a matter of getting enough DNA out of the sample. The DNA must accurately represent the different cell types and numbers of the microbial community to ensure effective diagnosis and treatment.
Additionally, the extraction method needs to be compatible with a variety of sample types (e.g. feces, urine, swabs, etc.), and ideally can be automated on robot platforms to streamline the processing of large quantities of samples. For this step, an extraction method like the ZymoBIOMICS 96 MagBead DNA Kit helps save time, and reduces errors and biases during the extraction step
Benefits of the ZymoBIOMICS 96 Magbead DNA Kit:
- Unbiased lysis of all microbes (bacteria, fungi, etc.)
- Consistent and streamlined processes of large quantities of samples, particularly using automation
- Compatibility with a variety of sample types, including those stored in DNA/RNA Shield stabilization reagent
-
Microbiome Standards and Mock Communities
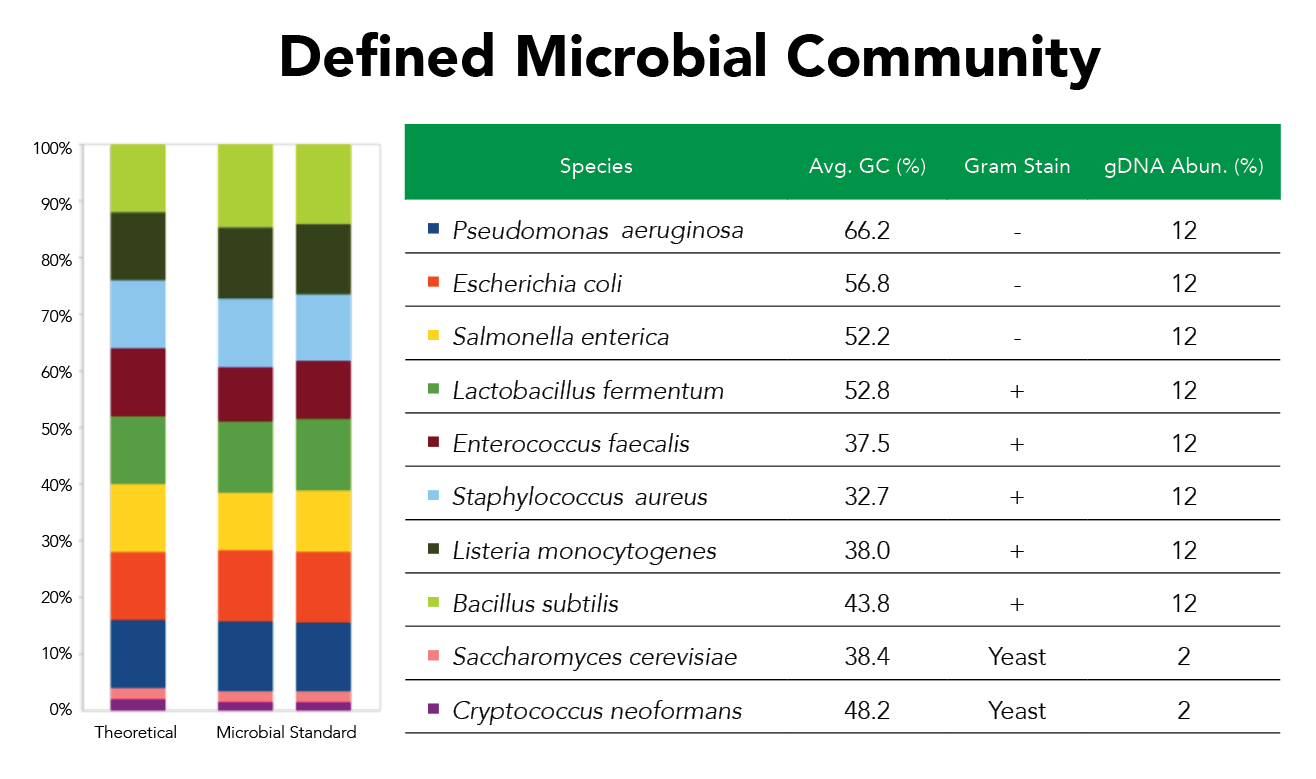
Figure 3. The ZymoBIOMICS Microbial Community Standard contains three easy-to-lyse bacteria, five tough-to-lyse bacteria, and two touch-to-lyse yeasts.
The use of well-defined and consistent Microbiome Standards and Mock Communities as positive controls is crucial for ensuring the accuracy and reliability of medical diagnostic reports for the MiDOG team. These controls serve as benchmarks that allow the lab to verify the performance of the sequencing process and the bioinformatics pipeline.
- Benchmarking Sequencing Accuracy
- Validation of Bioinformatics Pipelines
- Quality Control and Consistency
- Detection of Contamination
- Building Trust in Diagnostic Results
Incorporating these controls into an NGS-based diagnostic lab is a critical step in ensuring that every diagnostic report is accurate, reliable, and trustworthy.
-
NGS Library Preparation
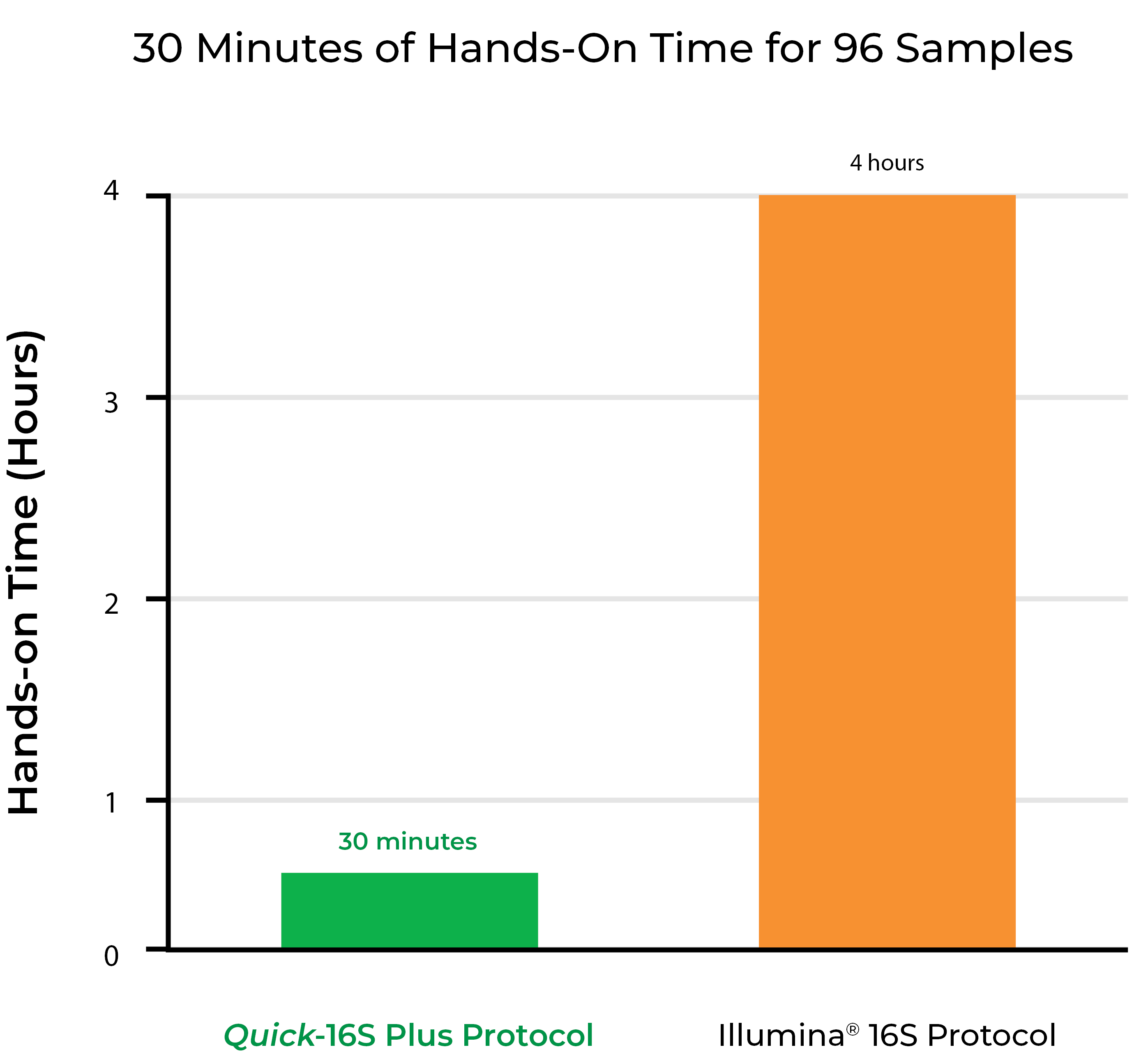
Figure 4. The Quick-16S Plus NGS Library Prep Kits dramatically reduce the hands-on time compared to the Illumina 16S Protocol so more samples can be processed in one day.
Finally, rapid turnaround is crucial for timely diagnoses, since it enables veterinarians to act swiftly in providing customized treatment. Usually, the library prep step can be the most challenging and time-consuming part of the workflow, especially since it requires much hand-on time. Thus, having a protocol that reduces hands on time, and can even be automated in a consistent way, allows the consistency and speed that are essential for accurate diagnostics across various species.
Benefits of the Quick-16S/ITS Library Kits:
- Streamlined protocol with only 30 minutes of hands-on time for 96 samples.
- 100% automation ready with only a single PCR step and no normalization needed.
- Real-time PCR enables absolute microbial copy number quantification
Want to Learn More ?
If you have any questions about the microbiome tools and solutions listed above, feel free to reach out to our Microbiome Scientists using the form below.