What is WGBS?
WGBS is a method to detect the DNA methylation status of nearly all the cytosines in the input sample. Genomic DNA is bisulfite converted, the library is prepared through a single- or double-stranded library preparation approach, and sequencing is done to generate single-base resolution data. As a result, it offers scientists a comprehensive profile of DNA methylation levels across the entire genome. The utilization of WGBS data enhances the likelihood of unveiling novel DNA methylation patterns through de novo discovery1,2.
Depending on the genome's size, WGBS might necessitate several hundreds of millions of sequencing reads to achieve sufficient depth for accurate methylation analysis. The advent of new sequencing technologies has substantially lowered the cost associated with WGBS.
Variations of the WGBS workflow have been developed to work with challenging sample types, such as cfDNA or FFPE DNA. These sample types are notorious for their low DNA integrity and/or limited availability. Commercially available kits like the Zymo-Seq Trio WGBS Library Kit have been developed with these hurdles in mind. This kit utilizes a single-stranded library preparation approach, and sequencing libraries can be made even from low input DNA or highly fragmented DNA.
While WGBS enables a detailed methylation analysis across the genome and is adaptable to a variety of challenging sample types, its high cost compared with alternative methods continues to serve as its main drawback. The benefits and challenges of using WGBS for genome-wide methylation assessment are summarized below.
Benefits of WBGS
-
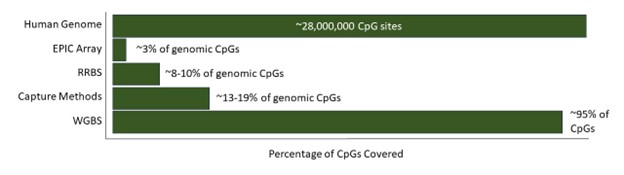
Comprehensive coverage of the majority of CpG sites, as well as non-CpG cytosines (CHG and CHH sites).
-
Can cover both high- and low-density CpG regions, ensuring thorough methylation analysis across the genome.
-
Bisulfite sequencing remains the gold standard for assessing methylation at single-base resolution.
-
Workflows are available for a variety of DNA inputs (i.e., low-input, FFPE DNA, cell-free DNA).
Challenges of WGBS
-
Despite improvements in cost efficiency, the cost of achieving whole-genome coverage with WGBS can still be prohibitive, especially for studies involving large sample sizes.
-
Cannot differentiate between 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC).
-
Can only detect methylation on cytosine residues.
WGBS vs. RRBS
The increased cost-effectiveness of WGBS positions it as the primary discovery platform for identifying markers in targeted clinical applications. After markers have been identified, follow-up studies can use targeted approaches (i.e., targeted bisulfite sequencing) to focus on specific loci and perform validation with increased sequencing depth. WGBS is a species -agnostic method that can be applied to samples from almost any species where DNA methylation on cytosine is of interest.
An alternative to WGBS is Reduced Representation Bisulfite Sequencing (RRBS), which is a more cost-effective approach for assessing DNA methylation in large-scale studies. However, RRBS relies on the restriction enzyme MspI to cut the genomic DNA at specific sequence motifs which are more common in CpG-dense regions. The choice of enzyme was based on the context in which methylated cytosines occur in the mammalian genome. For non-mammalian species, different enzymes may have to be validated for optimal coverage.
The choice between the two genome-wide methylation assays will depend on a variety of factors. These include, but are not limited to, scale of the study, the species of interest, the desired breadth of coverage, and the budget of the researcher.
Industry-Leading Solutions for DNA Methylation Studies
Zymo Research offers a comprehensive solution for DNA methylation profiling, streamlining your workflow from start to finish. Our industry-leading DNA purification technologies efficiently recover NGS-ready DNA from any sample type. This purified DNA seamlessly integrates with our bisulfite conversion and library preparation kits, simplifying your epigenomics research. Incorporating DNA methylation standards into your bisulfite sequencing workflow ensures precise validation of experimental conditions, delivering highly accurate and reliable NGS results. For labs looking to save time and resources, our Epigenome Sequencing Services offer a hassle-free way to uncover valuable insights from complex epigenetic patterns with ease.
With decades of experience in epigenetics, Zymo Research is uniquely positioned to support your projects using state-of-the-art products and services for DNA methylation analysis. From sample preparation to sequencing and reporting, Zymo Research scientists are available to support you every step of the way.
Citations
- Zhou, L.; Ng, H. K.; Drautz-Moses, D. I.; Schuster, S. C.; Beck, S.; Kim, C.; Chambers, J. C.; Loh, M. Systematic evaluation of library preparation methods and sequencing platforms for high-throughput whole genome bisulfite sequencing. Sci Rep 2019, 9 (1), 10383.
- Sun, Z.; Cunningham, J.; Slager, S.; Kocher, J. P. Base resolution methylome profiling: considerations in platform selection, data preprocessing and analysis. Epigenomics 2015, 7 (5), 813-828.
- Meissner A.; Gnirke A.; Bell G.W.; Ramsahoye B.; Lander E. S.; Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005 33 (18), 5868-77.