Successfully Added to Cart
Customers also bought...
-
 DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the...
DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the... -
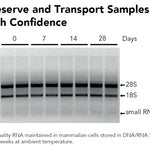
 DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
Highlights
- Automation-specific streamlined design for high-throughput bisulfite conversion of DNA for methylation analysis.
- Compatible with Illumina® Infinium™ MethylationEPIC, HTS iSelect® Methyl Custom, and Mouse Methylation arrays.
- Optimized buffer volumes for consistent automation performance.
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
Highlights
- Automation-specific streamlined design for high-throughput bisulfite conversion of DNA for methylation analysis.
- Compatible with Illumina® Infinium™ MethylationEPIC, HTS iSelect® Methyl Custom, and Mouse Methylation arrays.
- Optimized buffer volumes for consistent automation performance.
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
| Cat # | Name | Size | |
|---|---|---|---|
| D5032-1 | Lightning Conversion Reagent | 15 ml | |
| D5049-3 | M-Binding Buffer | 100 ml | |
| D5007-4 | M-Wash Buffer | 36 ml | |
| D5046-5 | L-Desulphonation Buffer | 80 ml | |
| D5049-6 | M-Elution Buffer | 50 ml | |
Description
Performance
Technical Specifications
| Binding Capacity | 1 µg total DNA per 5 µl MagBinding Beads |
|---|---|
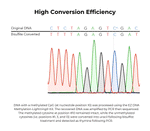
| Conversion Efficiency | > 99.5% of non-methylated C residues are converted to U; > 99.5% protection of methylated cytosines. |
| DNA Purity | A260/A280 & A260/A230 ≥ 1.8 |
| Input | 100 pg - 2 µg of DNA. For optimal results, the amount of input DNA should be from 200 to 500 ng. |
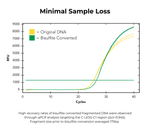
| Input Recovery | > 80% (≥ 100 bp) |
| Required Equipment | Thermal cycler with heated lid. |
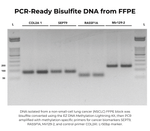
| Sample Source | Purified genomic DNA, endonuclease-digested DNA, linearized plasmid DNA, etc. DNA should be high-quality and RNA-free. |
| User Provided Materials | Ethanol (95 – 100%), non-skirted 96-well microplates, and 96-well microplate sealing films. See Recommendations for instrument hardware and labware requirements. |
Resources
Documents
FAQ
The EZ DNA Methylation-Lightning automation kit features optimized buffer volumes that allow for consistent high-throughput automation performance.
While we do not offer scripts for non-verified instruments, we can provide general technical support for any automation instrument.
Yes, we provide a guideline for users to create their own script in the protocol.
For best results, keep the method of quantification consistent before and after bisulfite treatment:
- If quantifying with a NanoDrop, use dsDNA settings (50 μg/ml for Ab260 = 1.0) before treatment and use RNA settings (40 μg/ml for Ab260 = 1.0) after treatment.
- If quantifying with Qubit, use a dsDNA assay before treatment and use a ssDNA assay after treatment.
Following bisulfite treatment, DNA will be single stranded with limited non-specific base pairing at room temperature. To visualize, run the converted DNA on an agarose gel then chill the gel on ice or in an ice bath for 30 minutes. This will force enough base-pairing to allow intercalation of the ethidium bromide for the DNA to be visible. If using a Bioanalyzer or TapeStation instrument, use RNA kits and reagents to visualize the converted DNA.
Converted DNA eluted in M-Elution Buffer can generally be stored at -20°C for 1-3 months. If longer term storage is necessary, we recommend storing at or below -70°C if possible. Bisulfite converted DNA is less stable than dsDNA; for best results, minimize freeze-thawing of converted DNA and use as soon as possible for downstream analysis.
Need help? Contact Us