Successfully Added to Cart
Updated Cart Total:
0
items
Subtotal:
$0.00 USD
Customers also bought...
-
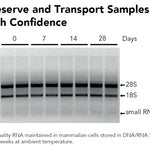 DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the...
DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the... -
 DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
Highlights
- Negative control for epigenetic experiments requiring DNA containing zero background levels of cytosine and adenosine methylation. Substrate for DNA methylation-sensitive restriction enzyme digestions.
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
Highlights
- Negative control for epigenetic experiments requiring DNA containing zero background levels of cytosine and adenosine methylation. Substrate for DNA methylation-sensitive restriction enzyme digestions.
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
Description
Performance
Technical Specifications
| Concentration | 250 ng/µl in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) |
|---|---|
| Genotype | ara-14 leuB6 fhuA31 lacY1 tsx78 glnV44 galK2 galT22 mcrA dcm-6 hisG4 rfbD1 R(zgb210::Tn10)TetS endA1 rpsL136 dam13::Tn9 xylA-5 mtl-1 thi-1 mcrB1 hsdR2 |
| Storage | ≤ -20°C |
Resources
Documents
FAQ
During NGS library prep, this standard can be spiked in to or run in parallel with experimental samples to monitor bisulfite conversion efficiency and/or workflow performance.
The reference genome of E. coli strain K-12 substrain MG1655 can be used for alignment and analysis. It can be accessed here.
Need help? Contact Us