Successfully Added to Cart
Customers also bought...
-
 DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the...
DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the... -
 DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
Format
ZymoBIOMICS DNA/RNA Miniprep Kit
Highlights
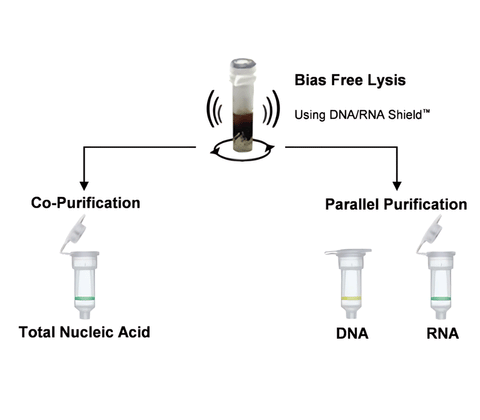
- Unbiased Lysis: Efficient and unbiased lysis of microbes including Grampositive/negative bacteria, fungi, protozoans, and viruses from any sample (feces, soil, plant, water, biofilms, swabs, saliva, body fluids, etc.)
- Ultra-Pure: DNA and Total RNA (including small/micro RNAs) is inhibitor-free and ready for qPCR and microbiome measurements using Next-Gen Sequencing.
- High Sensitivity: Increased detection limit of very low abundance organisms.
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
Format
ZymoBIOMICS DNA/RNA Miniprep Kit
Highlights
- Unbiased Lysis: Efficient and unbiased lysis of microbes including Grampositive/negative bacteria, fungi, protozoans, and viruses from any sample (feces, soil, plant, water, biofilms, swabs, saliva, body fluids, etc.)
- Ultra-Pure: DNA and Total RNA (including small/micro RNAs) is inhibitor-free and ready for qPCR and microbiome measurements using Next-Gen Sequencing.
- High Sensitivity: Increased detection limit of very low abundance organisms.
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
| Cat # | Name | Size | |
|---|---|---|---|
| E1010-1-4 | DNA Digestion Buffer | 4 mL | |
| C1058-50 | Zymo-Spin III-HRC Filters | 50 Pack | |
| C1001-50 | Collection Tubes | 50 Pack | |
| C1006-50-G | Zymo-Spin IIICG Columns | 50 Pack | |
| C1006-50-F | Spin-Away Filters | 50 Pack | |
| D7001-1-50 | DNA/RNA Lysis Buffer | 50 ml | |
| D7010-2-50 | DNA/RNA Prep Buffer | 50 ml | |
| D7010-3-24 | DNA/RNA Wash Buffer (Concentrate) | 24 ml | |
| R1100-50 | DNA/RNA Shield | 50 ml | |
| W1001-30 | DNase/RNase-Free Water | 30 ml | |
Description
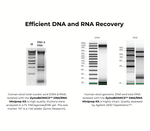
Performance
Technical Specifications
| Equipment | Microcentrifuge, vortex, cell disrupter (recommended) |
|---|---|
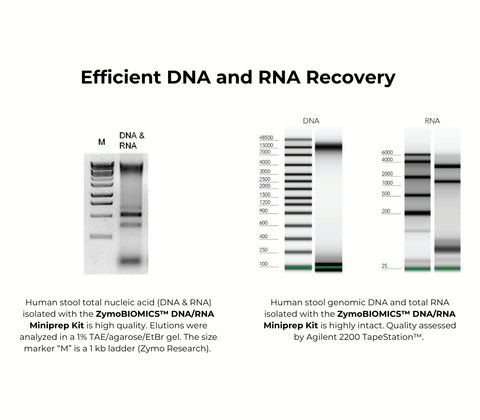
| Purity | DNA and RNA is ready for Next-Gen sequencing, RTPCR, microarray, hybridization, etc. A260/A280, A260/A230: >1.8. |
| Sample Source | Bacterial, fungal, protozoan, algae, viral, mitochondrial, and host RNA is efficiently isolated from ≤ 200 mg of mammalian feces, ≤ 250 mg soil, ≤ 200 mg plant/seed, 50-100 mg (wet weight) fungal bacterial cells, biofilms, and water. |
| Size Range | DNA and Total RNA ≥17 nt |
| Yield | 100 µg DNA/RNA (binding capacity), ≥50 µl (elution volume) |
Resources
Documents
FAQ
Environmental samples often contain inhibitors such as polyphenolics, humic/fulvic acids, tannins, melanins, etc. that affect downstream applications such as PCR. The Zymo-Spin III-HRC is designed to remove these PCR inhibitors and does not bind DNA/RNA. The Zymo-Spin III-HRC can be purchased separately as the OneStep PCR Inhibitor Kit (D6030).
Check sample type and input amount. Low biomass samples such as swabs, some soil samples, water, air are expected to have low yield. High biomass samples such as feces may cause overloading of the spin column. Using less input may improve yields.
Ensure sample is fully homogenized. Depending on bead beater used, the settings may have to be optimized for each sample. The homogenization time may be extended to see if yields improve.
Apply heated elution buffer (60-70° C) and allow elution buffer to incubate on the column for several minutes prior to elution.
Ensure that the DNA Elution Buffer is added directly to the column matrix.
Low A260/230 values can be due to handling issues (e.g. transfer of column during wash steps). To minimize reagent wash buffer carryover, centrifuge the column at max speed for 1 minute prior to elution.
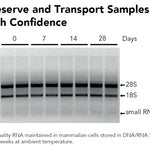
The chemistry extracts total RNA (including small RNAs and miRNAs) so an intense band at <200 nt size is within reason. For actual degraded RNA, make sure to check the sample collection procedures and storage conditions to ensure the initial sample is high quality. Degraded RNA can also occur if the bead-beating sample tube is overheated during homogenization. Reducing the time and intensity may improve performance.
Sample overloading can cause nucleic acids to precipitate and crash out of solution, reducing yields. Use less sample input to avoid precipitation.
This is a normal occurrence as DNA/RNA Shield contains detergents. To reduce foam, centrifuge at ≥ 12,000 x g in 1 minute intervals before transferring the supernatant.
We have validated our kits with both high-speed homogenizers and low-speed disruptors. We highly recommend users to optimize their bead beating conditions for their own instruments. We recommend using a 2 ml-tube adapter to ensure that the bead beating is efficient (do not use adapters made of foam). You can reference the Optimized Lysis Protocols table under documents for some recommendations.
This can happen on occasion due to transport or storage at lower temperatures. The reagent functionality is not affected; however, the precipitate can be resolved by heating the reagent to >37 °C.
Citations
Workflow Generator
Build Your Custom Microbiome Workflow
A great tool to help build or complete your workflow with our comprehensive microbiome solutions!
Need help? Contact Us