Successfully Added to Cart
Customers also bought...
-
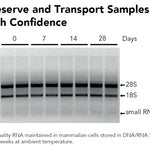 DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the...
DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the... -
 DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
Highlights
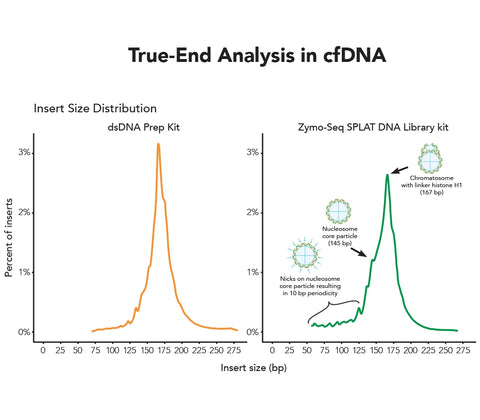
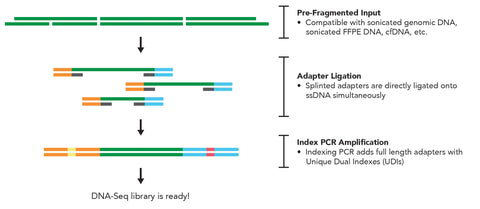
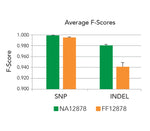
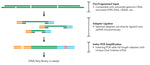
- Novel Solution for Fragmentomics Analysis: Powered by Splinted Ligation Adapter Tagging (SPLAT) technology, the kit precisely captures the true fragment ends of cell-free DNA, enabling accurate and high-fidelity analysis of fragmentation patterns.
- Versatile Sample Handling: The kit supports a wide range of sample types, including cell-free DNA (cfDNA), FFPE-derived DNA, and genomic DNA, while ensuring true end ligation for reliable and precise genomic data across diverse research applications.
- Efficient Workflow: The two-step workflow allows for the preparation of DNA samples into sequencing-ready libraries in as little as three hours, significantly speeding up the research process.
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
Highlights
- Novel Solution for Fragmentomics Analysis: Powered by Splinted Ligation Adapter Tagging (SPLAT) technology, the kit precisely captures the true fragment ends of cell-free DNA, enabling accurate and high-fidelity analysis of fragmentation patterns.
- Versatile Sample Handling: The kit supports a wide range of sample types, including cell-free DNA (cfDNA), FFPE-derived DNA, and genomic DNA, while ensuring true end ligation for reliable and precise genomic data across diverse research applications.
- Efficient Workflow: The two-step workflow allows for the preparation of DNA samples into sequencing-ready libraries in as little as three hours, significantly speeding up the research process.
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
| Cat # | Name | Size | |
|---|---|---|---|
| D3008 | Zymo-Seq UDI Primer Set (Indexes 1-12) | 12 indexes | |
| D4003-2-6 | DNA Wash Buffer (Concentrate) | 6 ml | |
| D3004-4-4 | DNA Elution Buffer | 4 ml | |
| W1001-1 | DNase/RNase-Free Water | 1 ml | |
Description
Performance
Technical Specifications
| Sample Input Type | Sonicated genomic DNA (average size of 300-600 bp), sonicated FFPE-derived DNA (average size of 300-600 bp), cfDNA, etc. |
|---|---|
| Minimum Input | 10 ng |
| Maximum Input | 500 ng |
| Input Quality | For optimal results, input DNA should be free of RNA contamination and enzymatic inhibitors with A260/A280 and A260/A230 values of ≥1.8. DNA with lower purity ratios (A260/A280 & A260/A230) should first be treated with the DNA Clean & Concentrator™ (Cat. No. D4013) prior to processing. DNA should be suspended in water, DNA Elution Buffer, TE buffer, or another similar low-salt buffer. |
| Equipment Required | Sonication device for DNA input >600 bp in average size, thermal cycler(s) with temperature-adjustable lids, microcentrifuge for 0.2 mL PCR tubes, magnetic stand for 0.2 mL PCR tubes (e.g., PCR Strip MagStand, Cat. No. 3DP-1002). |
| Total Processing Time | ~3 hours |
| Hands-On Time | ~1.5 hours |
| Library Storage | Libraries eluted in DNA Elution Buffer (provided) may be stored at ≤ 4°C overnight or ≤ -20°C for long-term storage. |
| Sequencing Platform Compatibility | Libraries are compatible with all Illumina® sequencing platforms. Please see Appendix D for sequencing considerations and recommendations. |
| Barcode Sequences | Available in Appendix C on pg. 14. Sequences are also available for download here (USA only) or by visiting the Documents section of the D3008 and D3096 product pages at www.zymoresearch.com. |
Resources
Documents
FAQ
It is possible to generate libraries with inputs lower than 10 ng. Typically, at lower inputs the percentage of adapter dimers increases in the final library. For best results, we recommend concentrating the sample input as much as possible to meet the 10 ng minimum input prior to using the kit. Samples can be concentrated using a DNA Clean & Concentrator™ Kit.
We recommend performing dilutions of adapters just before use in the ligation reaction for inputs <50 ng. For best results we do not recommend reusing previously diluted adapters.
Humidity and temperature vary between labs. Therefore, while we don’t recommend a specific amount of time to wait, we do recommend adding the elution buffer to the bead pellet as soon as the wet gloss dries and leaves the pellet matte in appearance. By holding the tube up to a bright light, this change is more easily seen in real time. Avoid adding the elution buffer when the bead pellet is still wet-looking, or after the pellet cracks as both may lead to a decrease in library yield. An example image showing different levels of “dryness” is shown here for your reference.

Each tube/well contains a pre-mixed forward and reverse primer set that contain a unique i5 and i7 index, respectively. The concentration of each UDI primer pre-mix is 5 µM (2.5 µM each primer).
Additional indexes 1-96 are sold separately as the Zymo-Seq UDI Primer Plate (Cat. No. D3096).
Each UDI primer set in the Zymo-Seq UDI Primer Set 1-12 (Cat. No. D3008) can be used more than once as long as the libraries with the same UDI set ARE NOT pooled and sequenced on the same sequencing lane. For more information regarding the UDI primer sets, please refer to Appendix C in the kit protocol or contact tech@zymoresearch.com.
This kit is compatible with any Illumina® sequencer. It is not directly compatible with Ion Torrent®, Oxford Nanopore®, PacBio®, or other non-Illumina platforms.
Libraries generated with this workflow are suitable for any read-length, but increased read lengths may require greater amounts of adapter trimming for the shorter library fragments. For most applications, 150 base paired-end (PE) reads are enough to generate substantial amounts of high-quality data for genome-wide coverage.
To estimate the necessary number of reads per library we recommend using the following Lander/Waterman equation:
Number of Reads = (Desired Coverage) × (Haploid Genome Length) / (Read Length)
Please note that this equation is helpful for estimating the number of reads necessary, however actual coverage may vary in practice. We also recommend using the Illumina® Sequencing Coverage Calculator to help determine the number of lanes/flow cells necessary for the desired sequencing instrument and parameters. Please contact Illumina® technical support for more information and recommendations.
The kit works best when DNA inputs >600 bp are fragmented using a physical sonication method. However, enzymatic methods have successfully been used, specifically with the dsDNA Shearase Plus (Cat. No. E2018). Please contact Zymo Research Technical Support at tech@zymoresearch.com for information on how to utilize the dsDNA Shearase Plus to perform enzymatic fragmentation before using this kit.
Need help? Contact Us