Successfully Added to Cart
Customers also bought...
-
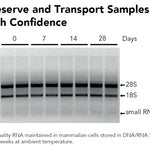 DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the...
DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the... -
 DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
Highlights
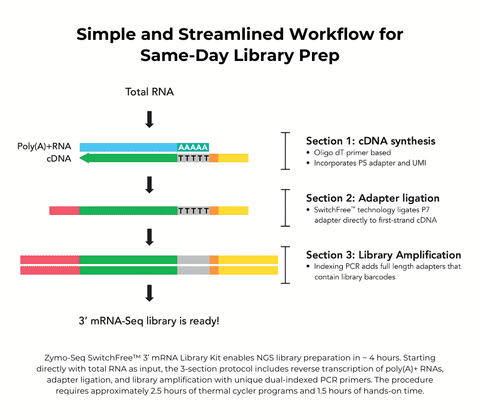
- Simplest protocol: RNA to library in less time with easy handling driven by the SwitchFree™ technology.
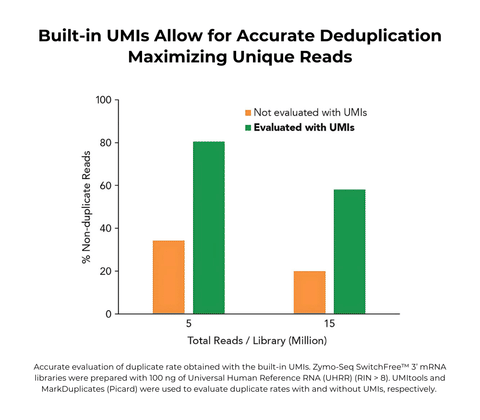
- High performance: Built-in Unique Molecular Identifiers (UMIs) allow for accurate deduplication maximizing unique reads.
- Low input compatible: Utilize as little as 10 ng total RNA without prior mRNA enrichment.
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
Highlights
- Simplest protocol: RNA to library in less time with easy handling driven by the SwitchFree™ technology.
- High performance: Built-in Unique Molecular Identifiers (UMIs) allow for accurate deduplication maximizing unique reads.
- Low input compatible: Utilize as little as 10 ng total RNA without prior mRNA enrichment.
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
| Cat # | Name | Size | |
|---|---|---|---|
| E2003 | ZymoTaq PreMix | (50 Rxns.) (2 x 625 µl) | |
| E2004 | ZymoTaq PreMix | (200 Rxns.) (8 x 625 µl) | |
| D4003-2-6 | DNA Wash Buffer (Concentrate) | 6 ml | |
| D4003-2-24 | DNA Wash Buffer (Concentrate) | 24 ml | |
| D4003-2-48 | DNA Wash Buffer (Concentrate) | 48 ml | |
| D3004-4-1 | DNA Elution Buffer | 1 ml | |
| D3004-4-4 | DNA Elution Buffer | 4 ml | |
| D3004-4-10 | DNA Elution Buffer | 10 ml | |
| D3004-4-16 | DNA Elution Buffer | 16 ml | |
| D3004-4-50 | DNA Elution Buffer | 50 ml | |
| W1001-1 | DNase/RNase-Free Water | 1 ml | |
| W1001-4 | DNase/RNase-Free Water | 4 ml | |
| W1001-6 | DNase/RNase-Free Water | 6 ml | |
| W1001-10 | DNase/RNase-Free Water | 10 ml | |
| W1001-30 | DNase/RNase-Free Water | 30 ml | |
| D3008 | Zymo-Seq UDI Primer Set (Indexes 1-12) | 12 indexes | |
| D3096 | Zymo-Seq UDI Primer Plate (Indexes 1-96) | 96 Indexes | |
| 3DP-1002 | PCR Strip MagStand | 1 unit | |
| P1005 | ZR-96 MagStand | 1 | |
Description
Performance
Technical Specifications
| Equipment Needed (user provided) | Thermal cycler with heated lid, magnetic stand for 0.2 mL PCR tubes, and microcentrifuge for 0.2 mL PCR tubes and 1.5 mL microcentrifuge tubes. |
|---|---|
| Input Quality | RNA should be free of DNA contamination and enzymatic inhibitors, with A260/A280 and A260/A230 ≥ 1.8. RNA with lower purity ratios (A260/A280 and A260/A230) should be treated with DNase I and purified with the RNA Clean & Concentrator™ (Cat. No. R1013) prior to processing. RNA should be suspended in water, TE, or a low-salt buffer.
|
| Library Storage | Libraries eluted in DNA Elution Buffer (provided) may be stored at ≤ 4°C overnight or ≤ -20°C for long-term storage. |
| Processing Time | ~4 hours |
| RNA Input | Compatible with 10 – 500 ng of total RNA. |
| Sequencing Platform Compatibility | Libraries are compatible with all Illumina® sequencing platforms except HiSeq® X. Libraries are compatible with the Element AVITI™ system. While technically possible, Illumina® originally limits the applications on HiSeq® X exclusively for whole-genome libraries. Please confirm with the sequencing service provider for acceptability and additional details if expecting to sequence Zymo-Seq SwitchFree™ 3′ mRNA libraries on HiSeq® X Series sequencers. |
Resources
Documents
FAQ
The kit should be compatible with purified total RNA from any organism where mature coding RNAs contain poly-A tails (e.g., eukaryotes).
Certain prokaryotes do have poly-A tails in their mRNA, therefore it may be technically possible to generate libraries from prokaryotic RNA. However, poly-A tails in prokaryotes may serve different functions than in eukaryotes (such as serving as a marker for mRNA degradation). Because the intrinsic functions of the poly-A tails differ between prokaryotic and eukaryotic organisms, we do not recommend SwitchFree 3′ mRNA-Seq to study prokaryotic organisms and cannot guarantee data quality if one proceeds with prokaryotic RNA.
Technically this kit can work with degraded RNA with low RIN scores, although the performance may be negatively impacted. Intrinsically, the method of 3′ mRNA library preparation may not capture protein coding transcripts as effectively from degraded inputs due to the likely damaged or missing poly-A tails.
If it is necessary to use degraded samples, we recommend increasing the input amount, amplifying with more PCR cycles and skipping Section 1, Step 4 of the protocol. Alternatively, for degraded RNA input consider using a total RNA library kit such as the Zymo-Seq RiboFree Total RNA Library Kit (R3000/R3003) which does not rely on intact poly-A tails.
It is possible to generate libraries with less than 10 ng of total RNA as input with this kit; however, the quality of such libraries is not guaranteed. Please contact tech@zymoresearch.com for recommendations and more details.
We have not directly tested this type of input with the SwitchFree kit, but in principle it should be possible to generate libraries using mRNA input. Please contact tech@zymoresearch.com for recommendations and more details.
DNA contamination will adversely impact the accuracy and sensitivity of quantitative measures for gene expression and differential gene expression analysis. Therefore, we recommend removing DNA contamination from the RNA input prior to performing the Zymo-Seq SwitchFree 3′ mRNA Library Kit workflow. For extracted RNA, we recommend using the RNA Clean & Concentrator-5 (R1013), which includes DNase I treatment and subsequent clean-up. This method has been validated for use with the Zymo-Seq SwitchFree 3′ mRNA Library Kit workflow.
Yes. All reagents to generate indexed, stranded cDNA libraries are included. These include the reagents for reverse transcription with built-in UMIs, adapter ligation, and library amplification with UDIs, as well as the magnetic beads for reaction cleanups.
The SwitchFree 3′ mRNA Library Kit generates libraries corresponding to mRNA (coding genes), whereas the RiboFree Total RNA Library Kit generates libraries that correspond to the entire transcriptome (both coding and non-coding genes). In addition, SwitchFree sequencing reads cover primarily the 3′ ends of transcripts, whereas RiboFree reads provide uniform 5′ to 3′ coverage. For more details, please see our blog "Considerations for Choosing Between Total RNA-Seq and 3' mRNA-Seq". Please contact tech@zymoresearch.com if you need further information.
Unique molecular identifiers (UMIs) are short sequences serving as molecular tags that enhance read deduplication and accuracy of gene expression quantification. Unique dual indexes (UDIs) are barcodes in the PCR primers that allow multiplexed libraries to be sequenced together. For more details, please see our blog “What Are UDIs and UMIs and Their Benefits in Next-Generation Sequencing?”. Please contact tech@zymoresearch.com if you need further information.
UMIs are built into the kit protocol. Therefore, they cannot be omitted during library preparation. However, their usage for bioinformatic analysis is optional. You can simply skip “Extracting UMIs” and proceed with bioinformatic analysis using Read 2 directly according to Appendix E: Bioinformatic Analysis in the protocol. For further information, please contact tech@zymoresearch.com.
This kit incorporates ion-based fragmentation of input RNA in Section 1: cDNA Synthesis along with other steps to generate cDNAs of appropriate sizes that are also enriched towards the 3’ end of transcripts with poly(A) tails.
If it is necessary to use degraded RNA samples as input for library preparation with this kit, we recommend increasing the input amount, amplifying with more PCR cycles, and skipping Section 1, Step 4 of the protocol.
Humidity and temperature vary between labs, so drying time can differ. We recommend starting with 5 minutes of air-dry time and adjusting as needed. Keep in mind that smaller volumes of beads will dry faster than larger volumes, so adjust the dry time accordingly when performing the Section 1, Section 2, and Section 3 bead cleanups. Please see the figure below for an example of over-, under-, and optimally (in the middle) dried beads.

The kit is designed to remove most adapter dimers. Occasionally, full-length adapter dimers may still remain. Please refer to Appendix C for recommendations on how to remove them.
Library yield can vary depending on factors such as the quality and quantity of the input RNA, and the PCR cycle number during library amplification. As an example, using 100 ng of Universal Human Reference RNA (RIN > 8.0) as input, the library yield is > 5 nmol/L when amplified with 15 PCR cycles and eluted to 20 μL of DNA Elution Buffer.
Besides being included in the 96-prep SwitchFree 3′ mRNA Library Kit (R3009), the UDI indexes 1-96 are also sold separately as the Zymo-Seq UDI Primer Plate (Cat. No. D3096).
For higher-plex applications requiring more than 96 unique dual indexes, we now offer the Zymo-Seq UDI Primer Plate (Indexes 1-96, 10-bp) and Zymo-Seq UDI Primer Plate (Indexes 97-192, 10-bp) (Cat. Nos. D3097 & D3098).
This kit is compatible with any Illumina® sequencer except for the HiSeq X Series. Illumina® originally limits the applications on HiSeq® X exclusively for whole-genome libraries. Please confirm with the sequencing service provider for acceptability and additional details if expecting to sequence Zymo-Seq SwitchFree™ 3′ mRNA libraries on HiSeq® X Series sequencers. It is not directly compatible with the Ion Torrent®, Oxford Nanopore®, PacBio® or other non-Illumina platforms.
For samples prone to dimer formation, in addition to using RNA of good quality and of as high input as possible (< 500 ng), we also recommend reducing the amount of PolyA R1 Reagent used. This will help further improve library quality as it reduces the likelihood of excess leftover primer. An example would be to use 3 μL of PolyA R1 Reagent instead of the 5 μL stated in Section 1 of the protocol. Supplement with 2 μL of DNase/RNase-Free Water to make up for the volume difference and proceed with the rest of the protocol as normal.

Reviews
"The primary strength of this kit was the simplicity of the workflow - by far, this is the easiest protocol I have encountered for RNA-seq library prep. I would recommend this to lab members as an entry point for RNA-seq. The stranded data and use of UMIs are two other major positives."
Jared T.
Miami University
"The kit was very straight-forward and easy to use."
Janice S.
Tarleton State University
Need help? Contact Us