Advancements In TRIzol® RNA Extraction
RNA extraction is among the first steps in studying gene expression with different methods, including northern blotting, transcriptome analysis, reverse transcription-PCR, and Next-Generation Sequencing (NGS). These techniques have become ubiquitous across myriad biomedical fields, from cancer to heart disease to neurodegeneration, positioning RNA purification as a foundational technique for these rapidly growing research areas.1-3
To support the remarkable pace and scale of these research fields, RNA extraction protocols need to be efficient and reliable. One method – acid-guanidinium-phenol-chloroform extraction4 – revolutionized the original protocol5 and became the “gold standard” for RNA extraction since 1987, remaining largely unchanged for the past few decades.
The key isolation reagent to this process, a monophasic solution of phenol and guanidinium thiocyanate, is commercially available under the brand names TRIzol® or Tri-Reagent®.6 These reagents lyse cells, release RNA, and prevent RNA degradation by inhibiting (most) RNases. They also separate lysate into different phases, enabling two other protocols (hence the “tri-“ prefix) using the interphase and organic phase for genomic DNA and protein purification, respectively.7
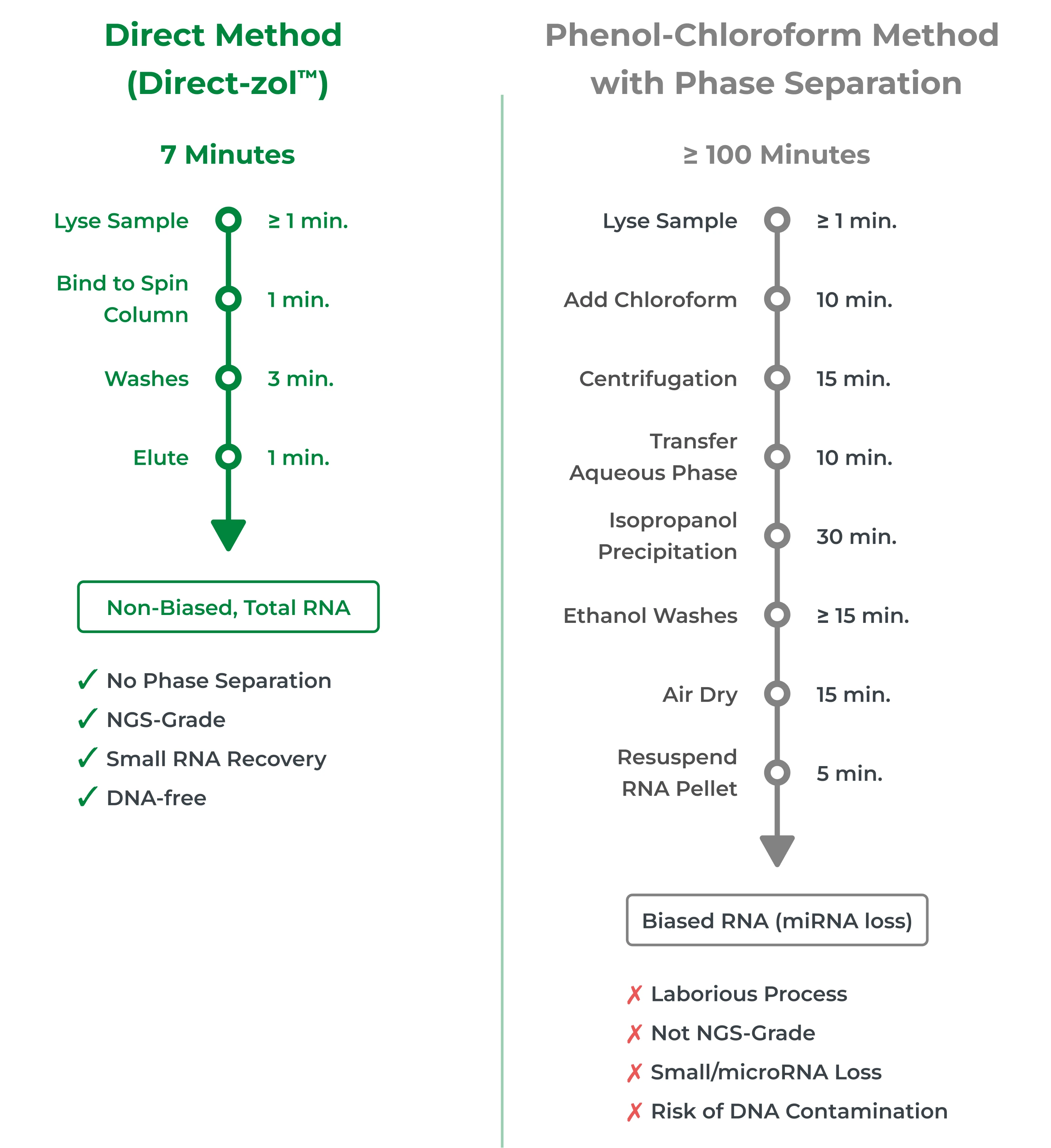
Nowadays, an even more streamlined method exists to remove the need for phase separation and reduces the ≥ 100-minute TRIzol® Phenol-Chloroform RNA extraction down to only 7 minutes. The Direct-zol™ method is the optimal solution for RNA extraction from TRIzol® due to its unsurpassed speed and ability to isolate high-yield, NGS-grade RNA, including small/miRNAs.

TRIzol® RNA Extraction Methods
TRIzol® RNA Extraction Protocols
TRIzol® RNA Extraction Protocol with Direct-zol Miniprep
MATERIALS
Equipment and Supplies
- Direct-zol™ RNA Miniprep Kit
- Centrifuge
- RNase-Free Microcentrifuge Tubes (1.5 – 2 mL)
Reagents
- Ethanol (95-100%)
PROTOCOL
Sample Preparation
Perform at room temperature and centrifugation at 10,000 - 16,000 x g for 1 minute.
- Cells:
- Remove growth media from culture dish, or collect cell suspension and centrifuge to discard the supernatant.
- Add 1 mL of TRIzol® per 1 x 105 – 1 x 107 cells or 0.1 mL TRIzol® per cm2 of culture surface area.
- Pipet mix lysate thoroughly to homogenize and lyse cells.
- Tough-to-Lyse Samples (e.g., Gram(+) bacteria, animal/plant tissue, etc.):
- Homogenized in ≥ 800 µl TRIzol® with ZR BashingBead™ Lysis Tubes (recommended), a mortar/pestle, dounce, syringe, tissue grinder, or bead beating with a high-speed homogenizer.
- To remove particulate debris from homogenized tissue, centrifuge and transfer the supernatant into a new RNase-free tube.
- Biological Liquids:
- Add ≥ 300 µl of TRIzol® to every 100 µl of liquid sample (e.g., cell suspensions, blood, plasma, serum, FACs, DNase I treated RNA etc.) and mix thoroughly.
- To remove particulate debris, centrifuge and transfer the supernatant into an RNase-free tube.
RNA Purification
Perform at room temperature and centrifugation at 10,000 - 16,000 x g for 30 seconds, unless specified.
- Add an equal volume ethanol (95-100%) to the sample lysed in TRIzol® and mix thoroughly.
- Transfer the mixture into a Zymo-Spin™ IICR Column in a Collection Tube and centrifuge.
- Transfer the column into a new collection tube and discard the flow-through.
- (Optional) Perform DNase I treatment with provided DNase I (see full protocol).
- Add 400 µl Direct-zol™ RNA PreWash to the column and centrifuge. Discard the flow-through.
- Repeat step 5.
- Add 700 µl RNA Wash Buffer to the column and centrifuge for 1 minute. Transfer the column carefully into an RNase-free tube.
- Add ≥ 25 µl of DNase/RNase-Free Water directly to the column matrix and centrifuge.
- The eluted RNA can be used immediately or stored frozen.
Phenol-Chloroform RNA Extraction (Phase Separation)
MATERIALS
Equipment and Supplies
- Centrifuge (capable of reaching up to 12,000 x g and down to 4°C)
- Water bath or heat block (55 – 60°C)
- RNase-Free Microcentrifuge Tubes (1.5 – 2 mL)
Reagents
- TRIzol®
- Chloroform
- Isopropanol
- Ethanol (75%)
- RNase-free water
PROTOCOL
Perform all steps at room temperature (20 – 25°C) unless specified.
Sample Preparation and Phase Separation
- Sample Lysis:
- Cells:
- Remove growth media from culture dish, or collect cell suspension and centrifuge to discard the supernatant.
- Add 1 mL of TRIzol® per 1 x 105 – 1 x 107 cells or 0.1 mL per cm2 of culture surface area.
- Pipet mix lysate thoroughly to homogenize and lyse cells.
- Tissues:
- For 50 – 100 mg of tissue, add 1 mL of TRIzol®.
- Homogenize using a homogenizer (e.g., ZR BashingBead Lysis Tubes)
- For samples with high fat content, centrifuge at 12,000 x g at 4°C for 5 – 10 minutes. Avoid the fatty layer and transfer the cleared lysate to a new microcentrifuge tube.
- Incubate the lysate at room temperature for 5 minutes to ensure complete lysis and dissociation of nucleoprotein complexes.
- Add 0.2 mL of chloroform for each mL of TRIzol® used for sample lysis. Cap the microcentrifuge tube, then thoroughly mix by shaking. Incubate at room temperature for 2 – 10 minutes.
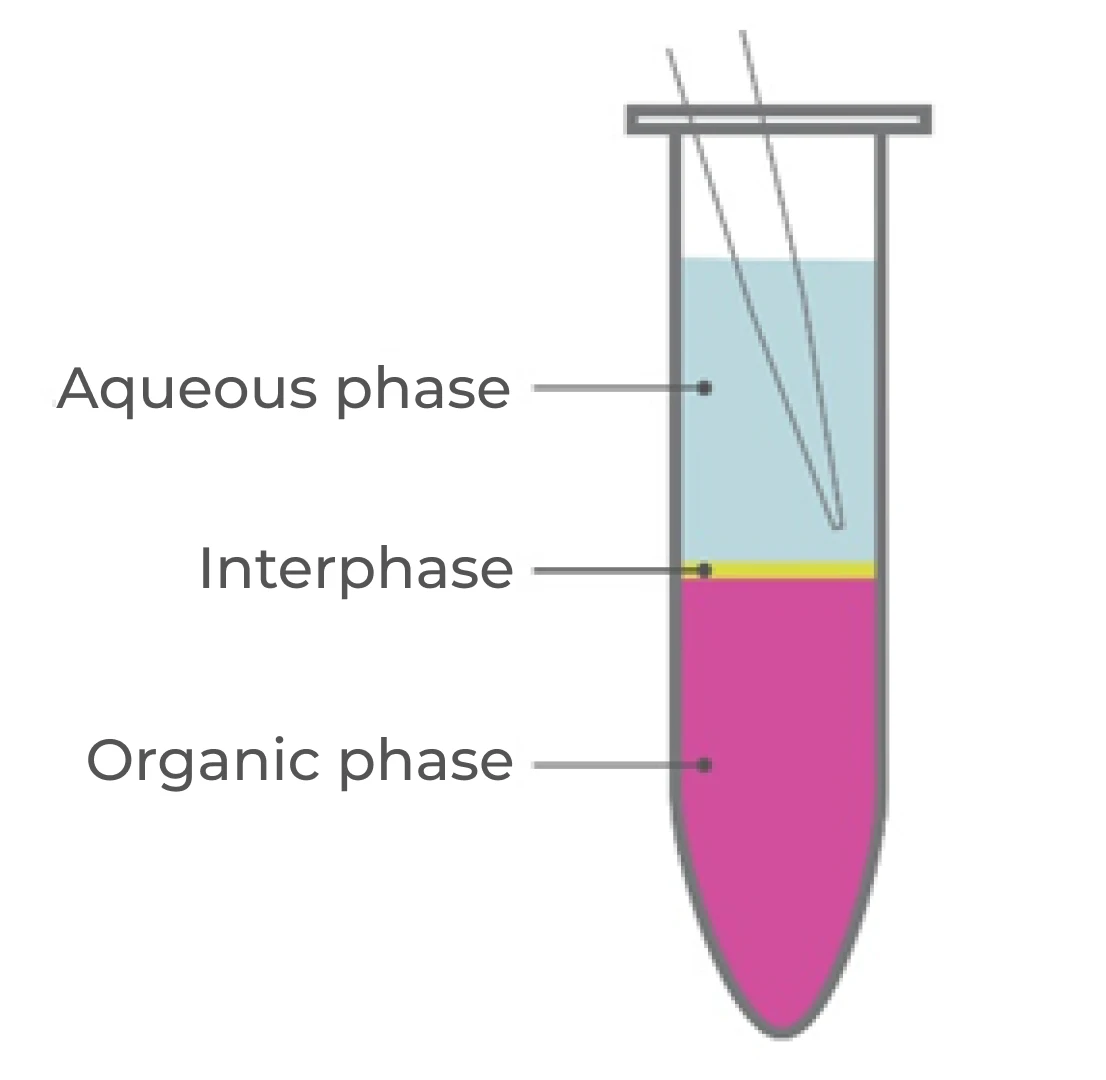
- Centrifuge at 12,000 x g at 4°C for 15 minutes, separating the mixture into three phases (Figure 1). Handle the tube gently to avoid disrupting the phases.
- Avoiding the interphase and organic phase, transfer the top aqueous phase to a new RNase-free tube through careful pipetting. Angle the tube at 45° for easier handling.
RNA Purification
Perform at room temperature and centrifugation at 10,000 - 16,000 x g for 30 seconds, unless specified.
- Add 0.5 mL of isopropanol to the transferred aqueous phase for each mL of TRIzol® used for lysis.
- Gently mix by inverting the tube several times. Incubate the mixture at 4°C for 10 minutes.
- Centrifuge the mixture at 12,000 x g at 4°C for ≥ 10 minutes.
- Discard the supernatant without disturbing the white pellet of RNA at the bottom of the tube.
- Add 1 mL of 75% ethanol to resuspend the RNA pellet for each 1 mL of TRIzol® used for lysis.
- Gently mix by inverting the tube several times.
- Centrifuge at 7,500 x g at 4°C for 5 – 10 minutes.
- Discard the supernatant without disturbing the RNA pellet at the bottom of the tube.
- Repeat the washing steps (step 5 – 8). Remove as much ethanol as possible through pipetting with a fine pipette tip (e.g., P10) without disturbing the RNA pellet.
- Air dry the RNA pellet for 5 – 15 minutes to evaporate residual ethanol. Avoid over-drying the pellet to allow easy solubilization of RNA.
- Resuspend the RNA pellet in RNase-free water. Incubating on a heat block at 55°C for 10 minutes can facilitate RNA solubilization.
- Proceed to downstream quantifications and applications, or store resuspended RNA at -70°C.
Tips For TRIzol® Phenol-Chloroform RNA Extraction
While performing TRIzol® RNA extraction with phase separation can seem straightforward, there are a few key challenges that many first-time extractors can face.
Tip 1: Set Aside the Right Amount of Time
While current TRIzol® Phenol-Chloroform RNA extraction protocols do not require 4 hours (as stated in the original 1987 paper), you will need about 2 hours, from sample lysis to resuspension of the dried RNA pellet. All the steps in the protocol are done manually, so the total extraction time largely depends on how many samples you have to process.
One of the major bottlenecks can be the centrifugation steps. Most microcentrifuges can hold between 20 to 30 samples, so if you are processing more samples than that, you may have to run your RNA extraction in batches, which can significantly increase your extraction time.
Tip 2: Beware of RNase
As you prepare your reagents, one key consideration is keeping them RNase-free. Despite what is published in the original 1987 paper, some RNases, like those in the RNase A family, are stabilized by disulfide bonds and remain active in TRIzol®.8 If you were to speak to any RNA extraction expert, they would tell you that these can wreak havoc on your purification and leave you with low quality, degraded RNA.
To avoid this, try using stabilization reagents or prepare a special RNase-free zone, where all reagents, tubes, pipette tips, and pipettes are RNase-free. In addition, your skin can be one of the sources of RNase contamination during an extraction, so try to minimize contact before, during, and after your RNA extraction.9
Tip 3: The Make-or-Break Step: Phase Separation
The major “make-or-break” step during RNA extraction is phase separation (Figure 1). It requires dexterity and careful pipetting to transfer as much of the aqueous (RNA) phase as possible without disturbing the DNA-rich interphase.
By disturbing the interphase, your purified RNA can be contaminated with genomic DNA, protein, and phenol, which can complicate quantification, reduce RNA quality, and inhibit downstream applications. This makes the phase separation step a major source of inconsistencies, such as impurities and yield variability across samples, resulting in inaccurate data analysis and low reproducibility. Furthermore, if you determine that you have genomic DNA contamination, you may have to follow up your RNA extraction with a DNase I digestion, which can add additional time to your overall workflow.
Tip 4: Understand the Bias: Low Inputs & Small RNAs
RNA extraction from TRIzol® relies on precipitation of RNA using isopropanol. With small amounts of RNA, precipitation is less efficient and can result in extremely low yields or no RNA pellet at all. Be sure to follow the manufacturer’s instructions and add the recommended input number of cells or tissue weight.
Precipitation is also inefficient for extracting small RNAs, like small/miRNAs. There can be biased results in the amount of small RNAs recovered.10 RNA extraction by phase separation was shown to selectively enrich for some species of small/miRNAs, leading to bias in downstream analysis. You can try using carriers, such as glycogen, to promote precipitation and improve your yield.11 Alternately, there are commercially available RNA purification kits specifically designed for small RNAs.
Extract RNA From TRIzol® In 7 Minutes
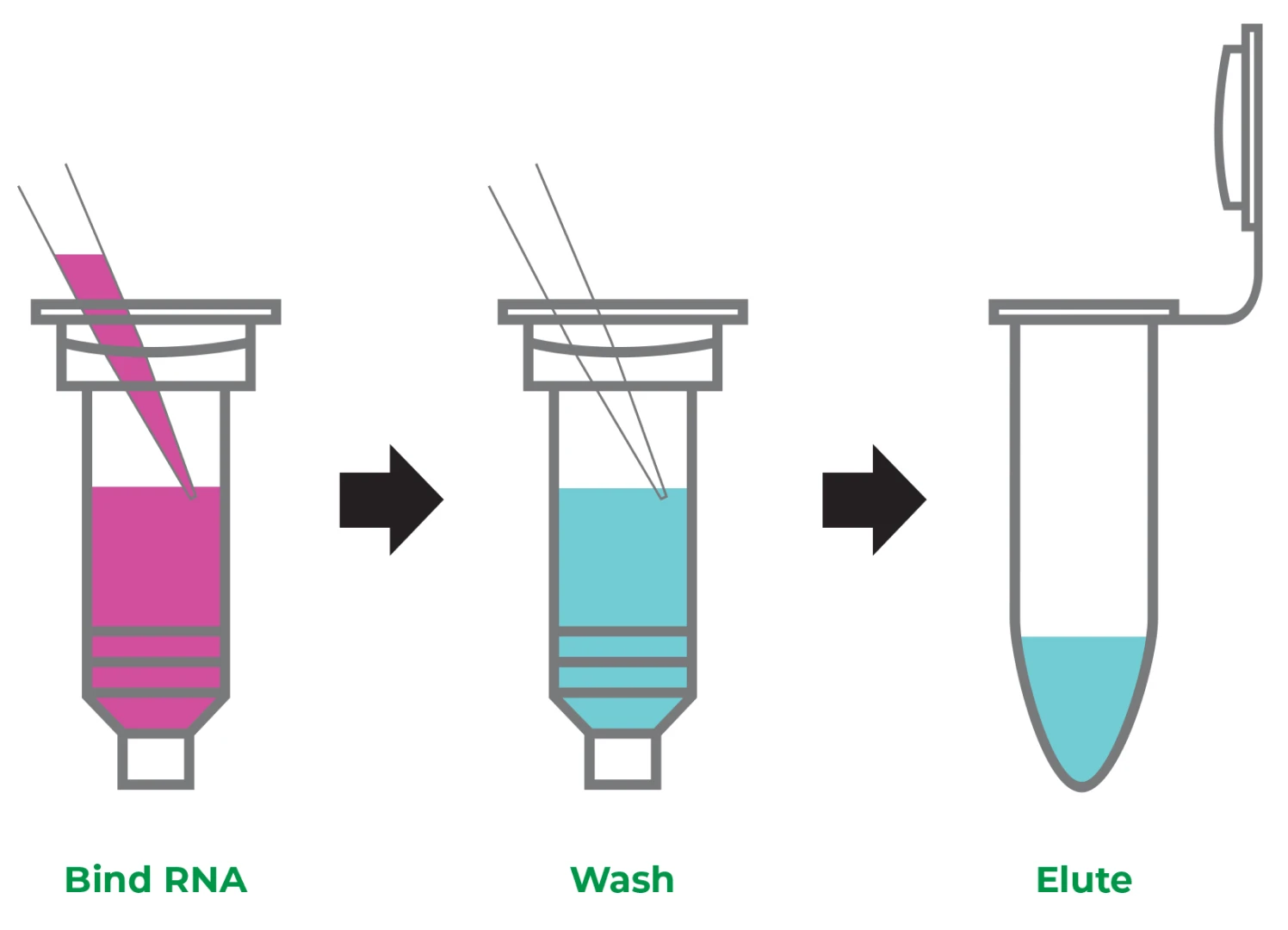
While TRIzol® RNA extraction is still considered the “gold standard,” there are a lot of limitations as mentioned above that make it unsuitable for your experiments. Zymo Research’s Direct-zol™ RNA Kits offer you a powerful alternative, reducing the entire RNA purification protocol from hours to just seven minutes. Starting with a sample resuspended in TRIzol®, there are just three simple steps: bind, wash, and elute RNA. First, add an equal volume of ethanol (1:1) to the lysed sample in TRIzol® and mix. Bind RNA to the silica-based column, wash the column, then elute RNA (Figure 2).
More than Speed
The Direct-zol™ RNA kit includes DNase I to rapidly remove contaminating DNA. It eliminates the need for chloroform (no phase separation) and concerns over phenol contamination or bias in small RNA recovery (Table 1).
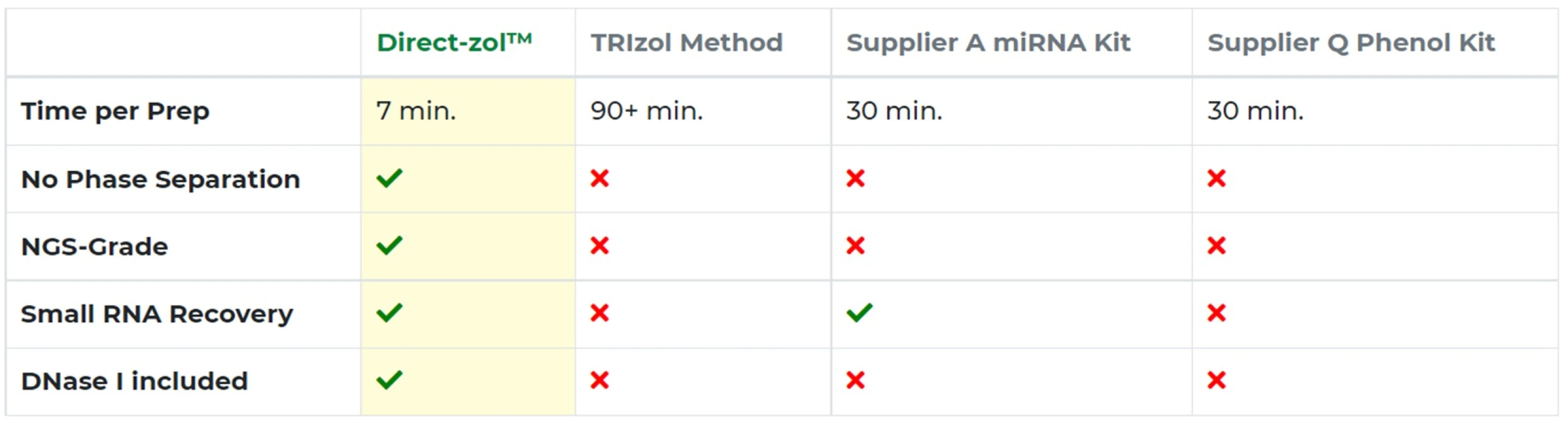
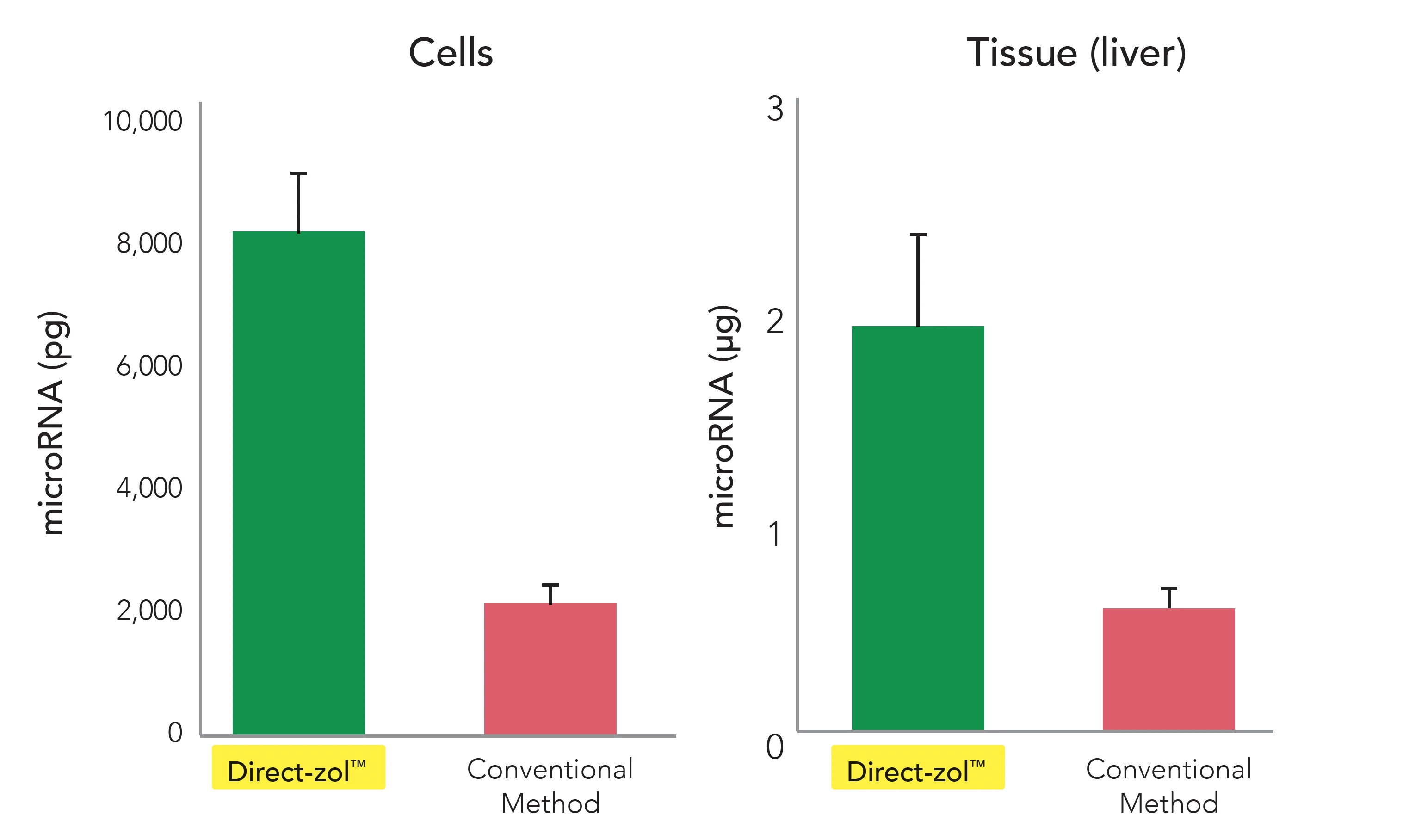
4X Small/Micro RNAs
At the end of the Direct-zol™ extraction process, the result is high-purity, high yield RNA – all the RNA. Direct-zol™ captures up to 4-times smaller/microRNAs than the traditional method and can deliver even picograms of RNA (Figure 3).
The Only Automated Solution for TRIzol® Samples
What is even more exciting is the possibility for high-throughput automation. Direct-zol™ has a bead-based solution which can be adapted for any automated platform, including Tecan, Hamilton, and Thermo’s KingFisher®. If you are interested, Zymo Research provides free automation scripts with technical support, as well as column-based sample kits for evaluation.
The future of diagnostics, drug discovery, vaccine development and more begins with high-quality RNA isolation, and Zymo Research has the tools to help your lab deliver the next big biomedical breakthrough.
Citations
- Khan P, Siddiqui JA, Lakshmanan I, et al. RNA-based therapies: A cog in the wheel of lung cancer defense. Mol Cancer. 2021;20(1):54.
- Lam YY, Keung W, Chan CH, et al. Single-Cell Transcriptomics of Engineered Cardiac Tissues From Patient-Specific Induced Pluripotent Stem Cell-Derived Cardiomyocytes Reveals Abnormal Developmental Trajectory and Intrinsic Contractile Defects in Hypoplastic Right Heart Syndrome. J Am Heart Assoc. 2020;9(20):e016528.
- Martier R, Konstantinova P. Gene Therapy for Neurodegenerative Diseases: Slowing Down the Ticking Clock. Front Neurosci. 2020;14:580179.
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162(1):156-159.
- Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18(24):5294-5299.
- Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat Protoc. 2006;1(2):581-585.
- Simões AE, Pereira DM, Amaral JD, et al. Efficient recovery of proteins from multiple source samples after TRIzol® or TRIzol®LS RNA extraction and long-term storage. BMC Genomics. 2013;14:181.
- Klink TA, Woycechowsky KJ, Taylor KM, Raines RT. Contribution of disulfide bonds to the conformational stability and catalytic activity of ribonuclease A. Eur J Biochem. 2000;267:566-572.
- Gupta SK, Haigh BJ, Griffin FJ, Wheeler TT. The mammalian secreted RNases: mechanisms of action in host defence. Innate Immun. 2013;19:86-97.
- Kim YK, et.al. Short structured RNAs with low GC content are selectively lost during extraction from a small number of cells. Mol Cell. 2012; 46(6); 893-85.
- Rio DC, Ares M Jr, Hannon GJ, Nilsen TW. Ethanol precipitation of RNA and the use of carriers. Cold Spring Harb Protoc. 2010;2010(6):pdb.prot5440.