What is PCR?
Polymerase chain reaction (PCR) is a reaction in which specific regions of DNA are amplified in vitro. It is one of the most widely utilized techniques in the science field because it is used for so many downstream molecular applications, including pathogen surveillance, cloning, in vitro diagnostics, DNA fingerprinting and genotyping.1 Furthermore, PCR is a critical step for targeted Next Generation Sequencing (NGS) because PCR can be used to enrich specific DNA fragments prior to sequencing. There are many different types of PCR reactions, such as reverse transcription-polymerase chain reaction (RT-PCR) and quantitative polymerase chain reaction (qPCR). The type of PCR reaction will depend on the type of nucleic acid template being used and if you want to quantify the amount of template present in the reaction. To learn more about the different kinds of PCR read our What is PCR and Different Types of PCR blog.
That said, all types of PCR require a mixture of polymerase enzyme, DNA or RNA template, a pair of primers, nucleotide bases, and co-factors, such as magnesium ions, to successfully amplify the target sequence. The components are then placed in a thermocycler machine where the changing temperature allows the DNA replication reaction to occur, as shown in Figure 1.
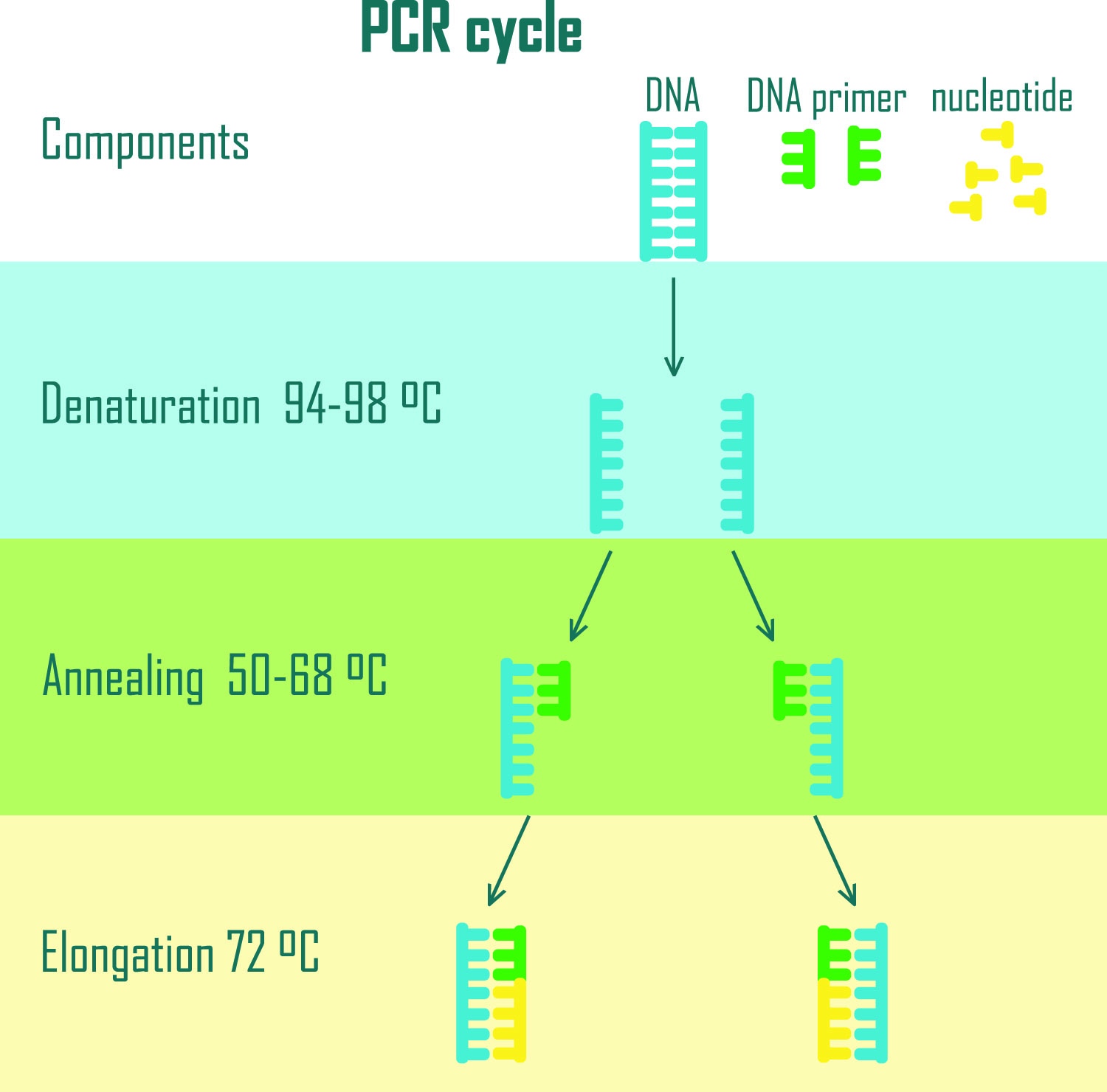
Figure 1. PCR cycle diagram showing in vitro DNA replication at different stages. The PCR mixture in the thermocycler is first exposed to high temperatures to separate DNA strands (Denaturation). The temperature is then decreased allowing the primers to anneal to the template DNA (Annealing), followed by a slight increase in temperature which allows the Taq Polymerase enzyme to elongate the strands (Elongation). This cycle is repeated to obtain millions of copy numbers of the desired DNA fragment.
As useful and revolutionary as PCR is, there are many ways it can fail due to the sensitivity of the reaction. One of the biggest obstacles to overcome when conducting sensitive reactions like NGS or PCR is the presence of PCR inhibitors.
What are PCR Inhibitors?
PCR inhibitors are organic and inorganic molecules that can interfere with PCR reactions.1 These molecules can range from simple salts like calcium or sodium to more complex ones like polyphenolics, detergents, polysaccharides, EDTA, etc.
PCR inhibitors can be introduced into the sample during sample preparation or co-extracted with the DNA or RNA template during purification. Environmental samples such as soil, feces, wastewater or plant samples are known to be rich in PCR inhibitors like polysaccharides or polyphenolics.1, 2 Other samples, such as fabrics, contain inhibitory dyes like indigo, and biological samples such as blood or tissue, can contain a number of highly inhibitory molecules, including hematin, collagen, or melanin.1
There are many ways in which these molecules can inhibit PCR. Some bind to the polymerase enzyme, preventing elongation, while others crosslink with the DNA template, preventing the strands from separating during the denaturation step.3 PCR inhibition can also occur when molecules, like EDTA or tannins, bind to cofactors of the DNA polymerase enzyme, such as Mg2+, which decreases the reaction rate or even completely inactivates the enzyme.3
Are PCR Inhibitors Impacting My Results?
A major consequence of an inhibited sample is that it can be interpreted as a false negative. In studies looking to detect the presence or absence of a certain DNA sequence, it may appear that the target is not present when it is possible it simply cannot be amplified because of high amounts of PCR inhibitors.
Currently, the simplest way to check for PCR inhibition is through dilution. When the sample is diluted, the inhibitors present in it are also diluted, so the inhibition is not as prominent (Figure 2). For context, in real time quantitative PCR there is a minimum threshold for fluorescence to be detected. The Ct (cycle threshold) value is the number of amplification cycles it takes for that threshold to be reached. A lower Ct value indicates there is more target DNA to begin with whereas a higher Ct value indicates there is less starting DNA because it takes more amplification cycles for the detection threshold to be reached.
In Figure 2a, a 1:10 dilution of an uninhibited sample results in a higher Ct value than the undiluted sample because it takes more cycles for the diluted sample to reach the minimum detection threshold. However, if inhibitors are present, than the 1:10 diluted sample will have a Ct equal to or lower than the undiluted sample, as shown in Figure 2b, because even if there is less sample there is also less PCR inhibition.
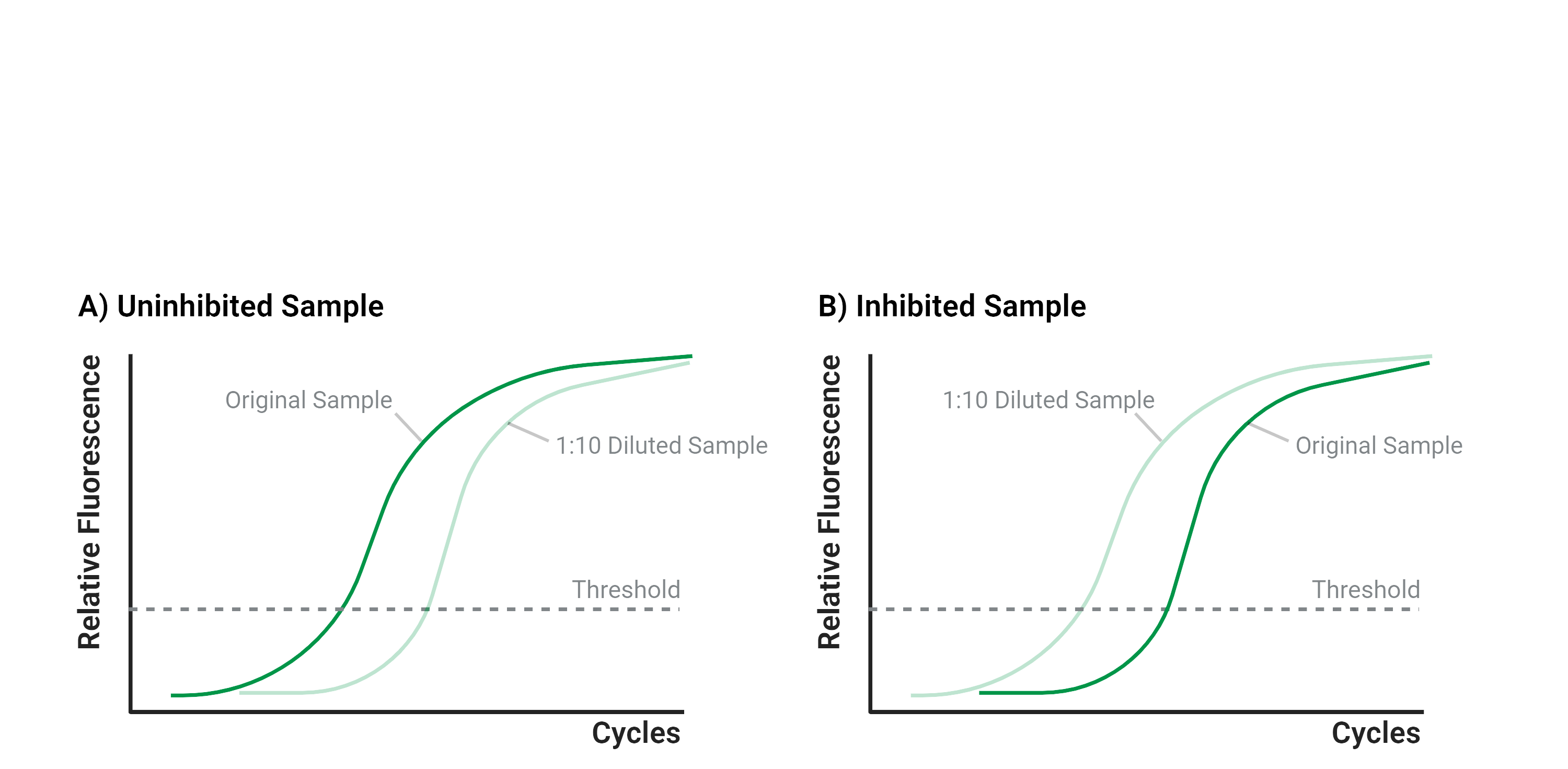
Figure 2. PCR amplification of an uninhibited sample compared to an inhibited sample. Figure 2a shows an uninhibited sample in which diluting the sample resulted in a higher Ct. Figure 2b shows an inhibited sample where the 1:10 dilution of the sample resulted in a lower Ct.
How to Remove PCR inhibitors
There are several ways to remove or decrease the effects of PCR inhibitors. Depending on the mechanism of inhibition, increasing DNA polymerase or reaction cofactors in the PCR reaction can decrease the effects of the inhibitors present in the sample. Sample dilution is also a common method to decrease the effects of inhibitors, however this leads to a decrease in the sensitivity of the PCR assay.
Some common methods for removal of PCR inhibitors include phenol-chloroform extraction, ion-exchange column chromatography, cation exchange columns, or CTAB based extraction.1 However, these traditional techniques for PCR inhibitor removal are tedious, time consuming, and can require harmful chemicals. In addition, oftentimes it is necessary to repeat these steps multiple times to completely remove inhibitors, which can lead to significant DNA and RNA loss. There are spin-column based solutions for cleaning up DNA and RNA, like Zymo Research’s DNA Clean & Concentrator-5, that are ideal for removing PCR inhibitors such as salts and detergents. However, if your sample type contains polyphenolic PCR inhibitors, they will still be co-purified with your nucleic acid. Zymo Research’s novel OneStep PCR Inhibitor Removal Kit is capable of completely removing polyphenolics from purified DNA & RNA in less than 5 minutes without the need for harmful chemicals like chloroform. The kit is equipped with a unique column matrix that binds common polyphenolic PCR inhibitors such as humic/fulvic acids, tannins, and melanin. Extracted DNA or RNA is passed through the column using one simple centrifugation step, minimizing nucleic acid loss. This exclusive technology is already included in a number of Zymo Research’s extraction kits, such as the Quick-DNA Fecal/Soil Microbe Kits and the Quick-RNA Plant Kit, ensuring recovered DNA or RNA is ready for all sensitive downstream applications (Figure 3).
| Extraction Kits with OneStep™ PCR Inhibitor Removal Technology Included | Cat. No. |
|---|---|
| ZymoBIOMICS RNA Kit | R2001 |
| ZymoBIOMICS DNA/RNA Kit | R2002 |
| ZymoBIOMICS DNA Kits | D4300, D4301, D4303, D4304, D4307, D4309 |
| Quick-DNA Fecal/Soil Microbe Kits | D6010, D6011, D6012, D6110 |
| Quick-DNA Plant/Seed Kit | D6020, D6021 |
| Quick-RNA Fecal/Soil Microbe Kit | R2040 |
| Quick-RNA Plant Kit | R2024 |
| Zymo Environ Water RNA Kit | R2042 |
Achieve PCR Ready Nucleic Acid Every Time
PCR is a globally used technique that is essential for applications like cancer diagnostics, metagenomics, GMO crop development, and pathogen surveillance in wastewater. No matter the application, highly accurate PCR results are essential for success and can be achieved by completely removing PCR inhibitors from extracted DNA or RNA. Utilize Zymo Research’s OneStep PCR Inhibitor Removal Kit to ensure your PCR and NGS results are accurate every time.
Citations
- Schrader, C., Schielke, A., Ellerbroek, L., & Johne, R. (2012). PCR inhibitors - occurrence, properties and removal. Journal of applied microbiology, 113(5), 1014–1026.
- Sidstedt, M., Rådström, P., & Hedman, J. (2020). PCR inhibition in qPCR, dPCR and MPS-mechanisms and solutions. Analytical and bioanalytical chemistry, 412(9), 2009–2023.
- Opel, K. L., Chung, D., & McCord, B. R. (2010). A study of PCR inhibition mechanisms using real time PCR. Journal of forensic sciences, 55(1), 25–33.